Abstract
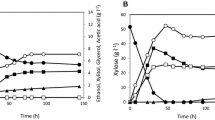
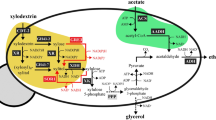
A recombinant Saccharomyces cerevisiae strain transformed with xylose reductase (XR) and xylitol dehydrogenase (XDH) genes from Pichia stipitis has the ability to convert xylose to ethanol together with the unfavorable excretion of xylitol, which may be due to cofactor imbalance between NADPH-preferring XR and NAD+-dependent XDH. To reduce xylitol formation, we have already generated several XDH mutants with a reversal of coenzyme specificity toward NADP+. In this study, we constructed a set of recombinant S. cerevisiae strains with xylose-fermenting ability, including protein-engineered NADP+-dependent XDH-expressing strains. The most positive effect on xylose-to-ethanol fermentation was found by using a strain named MA-N5, constructed by chromosomal integration of the gene for NADP+-dependent XDH along with XR and endogenous xylulokinase genes. The MA-N5 strain had an increase in ethanol production and decrease in xylitol excretion compared with the reference strain expressing wild-type XDH when fermenting not only xylose but also mixed sugars containing glucose and xylose. Furthermore, the MA-N5 strain produced ethanol with a high yield of 0.49 g of ethanol/g of total consumed sugars in the nonsulfuric acid hydrolysate of wood chips. The results demonstrate that glucose and xylose present in the lignocellulosic hydrolysate can be efficiently fermented by this redox-engineered strain.





Similar content being viewed by others
References
Amore R, Kötter P, Kuster C, Ciriacy M, Hollenberg CP (1991) Cloning and expression in Saccharomyces cerevisiae of the NAD(P)H-dependent xylose reductase-encoding gene (xyl1) from the xylose-assimilating yeast Pichia stipitis. Gene 109:89–97
Aristidou A, Penttilä M (2000) Metabolic engineering applications to renewable resource utilization. Curr Opin Biotechnol 11:187–198
Bruinenberg PM, de Bot PHM, van Dijken JP, Scheffers WA (1983) The role of redox balances in the anaerobic fermentation of xylose by yeasts. Eur J Appl Microbiol Biotechnol 18:287–292
Chu BC, Lee H (2007) Genetic improvement of Saccharomyces cerevisiae for xylose fermentation. Biotechnol Adv 25:425–441
Deng XX, Ho NW (1990) Xylulokinase activity in various yeasts including Saccharomyces cerevisiae containing the cloned xylulokinase gene. Appl Biochem Biotechnol 24–25:193–199
Eliasson A, Christensson C, Wahlbom CF, Hahn-Hägerdal B (2000) Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol 66:3381–3386
Eliasson A, Hofmeyr J-HS, Pedler S, Hahn-Hägerdal B (2001) The xylose reductase/xylitol dehydrogenase/xylulokinase ratio affects product formation in recombinant xylose-utilising Saccharomyces cerevisiae. Enzyme Microb Technol 29:288–297
Futcher AB, Cox BS (1984) Copy number and the stability of 2-μm circle-based artificial plasmids of Saccharomyces cerevisiae. J Bacteriol 157:283–290
Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20:1425
Gong CS, Cao NJ, Du J, Tsao GT (1999) Ethanol production from renewable resources. Adv Biochem Eng Biotechnol 65:207–241
Hahn-Hägerdal B, Wahlbom CF, Gárdonyi M, van Zyl WH, Cordero Otero RR, Jönsson LJ (2001) Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. Adv Biochem Eng Biotechnol 73:53–84
Hahn-Hägerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund MF (2007) Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv Biochem Eng Biotechnol 108:147–177
Hamacher T, Becker J, Gárdonyi M, Hahn-Hägerdal B, Boles E (2002) Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 148:2783–2788
Ho NW, Chen Z, Brainard AP (1998) Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol 64:1852–1859
Inoue H, Yano S, Endo T, Sakaki T, Sawayama S (2008) Combining hot-compressed water and ball milling pretreatments to improve the efficiency of the enzymatic hydrolysis of eucalyptus. Biotechnol Biofuels 1:2
Jeffries TW (2006) Engineering yeasts for xylose metabolism. Curr Opin Biotechnol 17:320–326
Jeffries TW, Jin YS (2004) Metabolic engineering for improved fermentation of pentoses by yeasts. Appl Microbiol Biotechnol 63:495–509
Jeffries TW, Grigoriev IV, Grimwood J, Laplaza JM, Aerts A, Salamov A, Schmutz J, Lindquist E, Dehal P, Shapiro H, Jin YS, Passoth V, Richardson PM (2007) Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat Biotechnol 25:319–326
Jeppsson M, Johansson B, Hahn-Hägerdal B, Gorwa-Grauslund MF (2002) Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose. Appl Environ Microbiol 68:1604–1609
Jeppsson M, Träff K, Johansson B, Hahn-Hägerdal B, Gorwa-Grauslund MF (2003) Effect of enhanced xylose reductase activity on xylose consumption and product distribution in xylose-fermenting recombinant Saccharomyces cerevisiae. FEMS Yeast Res 3:167–175
Jeppsson M, Bengtsson O, Franke K, Lee H, Hahn-Hägerdal B, Gorwa-Grauslund MF (2006) The expression of a Pichia stipitis xylose reductase mutant with higher K M for NADPH increases ethanol production from xylose recombinant Saccharomyces cerevisiae. Biotechnol Bioeng 93:665–673
Jin YS, Jones S, Shi NQ, Jeffries TW (2002) Molecular cloning of XYL3 (D-xylulokinase) from Pichia stipitis and characterization of its physiological function. Appl Environ Microbiol 68:1232–1239
Jin YS, Ni H, Laplaza JM, Jeffries TW (2003) Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate D-xylulokinase activity. Appl Environ Microbiol 69:495–503
Johansson B, Christensson C, Hobley T, Hahn-Hägerdal B (2001) Xylulokinase overexpression in two strains of Saccharomyces cerevisiae also expressing xylose reductase and xylitol dehydrogenase and its effect on fermentation of xylose and lignocellulosic hydrolysate. Appl Environ Microbiol 67:4249–4255
Katahira S, Mizuike A, Fukuda H, Kondo A (2006) Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Appl Microbiol Biotechnol 72:1136–1143
Kavanagh KL, Klimacek M, Nidetzky B, Wilson DK (2002) The structure of apo and holo forms of xylose reductase, a dimeric aldo-keto reductase from Candida tenuis. Biochemistry 41:8785–8795
Kavanagh KL, Klimacek M, Nidetzky B, Wilson DK (2003) Structure of xylose reductase bound to NAD+ and the basis for single and dual co-substrate specificity in family 2 aldo-keto reductases. Biochem J 373:319–326
Kostrzynska M, Sopher CR, Lee H (1998) Mutational analysis of the role of the conserved lysine-270 in the Pichia stipitis xylose reductase. FEMS Microbiol Lett 159:107–112
Kötter P, Ciriacy M (1993) Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 38:776–783
Lee J (1997) Biological conversion of lignocellulosic biomass to ethanol. J Biotechnol 56:1–24
Leitgeb S, Petschacher B, Wilson DK, Nidetzky B (2005) Fine tuning of coenzyme specificity in family 2 aldo-keto reductases revealed by crystal structures of the Lys-274→Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NAD+ and NADP+. FEBS Lett 579:763–767
Lin S, Tanaka S (2006) Ethanol fermentation from biomass resources: current state and prospects. Appl Microbiol Biotechnol 69:627–642
Matsushika A, Watanabe S, Kodaki T, Makino K, Sawayama S (2008) Bioethanol production from xylose by recombinant Saccharomyces cerevisiae expressing xylose reductase, NADP+-dependent xylitol dehydrogenase, and xylulokinase. J Biosci Bioeng 105:296–299
Meinander NQ, Hahn-Hägerdal B (1997) Fed-batch xylitol production with two recombinant Saccharomyces cerevisiae strains expressing XYL1 at different levels, using glucose as a cosubstrate: a comparison of production parameters and strain stability. Biotechnol Bioeng 54:391–399
Meinander NQ, Boels I, Hahn-Hägerdal B (1999) Fermentation of xylose/glucose mixtures by metabolically engineered Saccharomyces cerevisiae strains expressing XYL1 and XYL2 from Pichia stipitis with and without overexpression of TAL1. Bioresour Technol 68:79–87
Olsson L, Hahn-Hägerdal B (1993) Fermentative performance of bacteria and yeast in lignocellulose hydrolysates. Process Biochem 28:249–257
Olsson L, Nielsen J (2000) The role of metabolic engineering in the improvement of Saccharomyces cerevisiae: utilization of industrial media. Enzyme Microb Tech 26:785–792
Petschacher B, Nidetzky B (2005) Engineering Candida tenuis xylose reductase for improved utilization of NADH: antagonistic effects of multiple side chain replacements and performance of site-directed mutants under simulated in vivo conditions. Appl Environ Microbiol 71:6390–6393
Petschacher B, Nidetzky B (2008) Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae. Microb Cell Fact 7:9
Petschacher B, Leitgeb S, Kavanagh KL, Wilson DK, Nidetzky B (2005) The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and X-ray crystallography. Biochem J 385:75–83
Richard P, Toivari MH, Penttilä M (1999) Evidence that the gene YLR070c of Saccharomyces cerevisiae encodes a xylitol dehydrogenase. FEBS Lett 457:135–138
Richard P, Toivari MH, Penttilä M (2000) The role of xylulokinase in Saccharomyces cerevisiae xylulose catabolism. FEMS Microbiol Lett 190:39–43
Rizzi M, Harwart K, Erlemann P, Bui-Thanh NA, Dellweg H (1989) Purification and properties of the NAD+-xylitol-dehydrogenase from the yeast Pichia stipitis. J Ferment Bioeng 67:20–24
Shamanna DK, Sanderson KE (1979) Uptake and catabolism of D-xylose in Salmonella typhimurium LT2. J Bacteriol 139:64–70
Slininger PJ, Bothast RJ, Van Cauwenberge JE, Kurtzman CP (1982) Conversion of D-xylose to ethanol by the yeast Pachysolen tannophilus. Biotechnol Bioeng 24:371–384
Smiley KL, Bolen PL (1982) Demonstration of D-xylose reductase and D-xylitol dehydrogenase in Pachysolen tannophilus. Biotechnol Lett 4:607–610
Sonderegger M, Jeppsson M, Larsson C, Gorwa-Grauslund MF, Boles E, Olsson L, Spencer-Martins I, Hahn-Hägerdal B, Sauer U (2004) Fermentation performance of engineered and evolved xylose-fermenting Saccharomyces cerevisiae strains. Biotechnol Bioeng 87:90–98
Toivari MH, Aristidou A, Ruohonen L, Penttilä M (2001) Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae: importance of xylulokinase (XKS1) and oxygen availability. Metab Eng 3:236–249
Träff KL, Jönsson LJ, Hahn-Hägerdal B (2002) Putative xylose and arabinose reductases in Saccharomyces cerevisiae. Yeast 19:1233–1241
Verduyn C, Van Kleef R, Frank J, Schreuder H, Van Dijken JP, Scheffers WA (1985) Properties of the NAD(P)H-dependent xylose reductase from the xylose-fermenting yeast Pichia stipitis. Biochem J 226:669–677
Walfridsson M, Hallborn J, Penttilä M, Keränen S, Hahn-Hägerdal B (1995) Xylose-metabolizing Saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl Environ Microbiol 61:4184–4190
Walfridsson M, Anderlund M, Bao X, Hahn-Hägerdal B (1997) Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilisation. Appl Microbiol Biotechnol 48:218–224
Watanabe S, Kodaki T, Makino K (2005) Complete reversal of coenzyme specificity of xylitol dehydrogenase and increase of thermostability by the introduction of structural zinc. J Biol Chem 280:10340–10349
Watanabe S, Saleh AA, Pack SP, Annaluru N, Kodaki T, Makino K (2007) Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein engineered NADP+-dependent xylitol dehydrogenase. J Biotechnol 130:316–319
Yang VW, Jeffries TW (1997) Regulation of phosphotransferases in glucose- and xylose-fermenting yeasts. Appl Biochem Biotechnol 63–65:97–108
Zhang Z, Moo-Young M, Chisti Y (1996) Plasmid stability in recombinant Saccharomyces cerevisiae. Biotechnol Adv 14:401–435
Acknowledgments
We thank Ms. Maiko Kato for her technical support. This study was supported by the New Energy and Industrial Technology Development Organization (NEDO), Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Matsushika, A., Watanabe, S., Kodaki, T. et al. Expression of protein engineered NADP+-dependent xylitol dehydrogenase increases ethanol production from xylose in recombinant Saccharomyces cerevisiae . Appl Microbiol Biotechnol 81, 243–255 (2008). https://doi.org/10.1007/s00253-008-1649-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1649-1