Abstract
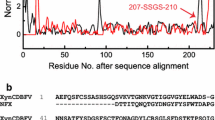
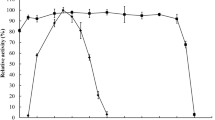
The extreme process condition of high temperature and high alkali limits the applications of most of natural xylanases in pulp and paper industry. Recently, various methods of protein engineering have been used to improve the thermal and alkalic tolerance of xylanases. In this work, directed evolution and site-directed mutagenesis were performed to obtain a mutant xylanase improved both on alkali stability and thermostability from the native Paenibacillus campinasensis Family-11 xylanase (XynG1-1). Mutant XynG1-1B43 (V90R/P172H) with two units increased in the optimum pH (pH 7.0–pH 9.0) and significant improvement on alkali stability was selected from the second round of epPCR library. And the further thermoduric mutant XynG1-1B43cc16 (V90R/P172H/T84C-T182C/D16Y) with 10 °C increased in the optimum temperature (60–70 °C) was then obtained by introducing a disulfide bridge (T84C-T182C) and a single amino acid substitution (D16Y) to XynG1-1B43 using site-directed mutagenesis. XynG1-1B43cc16 also showed higher thermostability and catalytic efficiency (k cat /K m ) than that of wild-type (XynG1-1) and XynG1-1B43. The attractive improved properties make XynG1-1B43cc16 more suitable for bioleaching of cotton stalk pulp under the extreme process condition of high temperature (70 °C) and high alkali (pH 9.0).






Similar content being viewed by others
References
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a Web-based environment for protein structure homology modelling. Bioinformatics 22(2):195–201
Bailey MJ, Biely P, Poutanen K (1992) Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol 23(3):257–270
Balaa BA, Brijs K, Gebruers K, Vandenhaute J, Wouters J, Housen I (2009) Xylanase XYL1p from Scytalidium acidophilum: site-directed mutagenesis and acidophilic adaptation. Bioresour Technol 100(24):6465–6471
Beg QK, Kapoor M, Mahajan L, Hoondal GS (2001) Microbial xylanases and their industrial applications: a review. Appl Microbiol Biotechnol 56(3–4):326–338
Birijlall N, Manimaran A, Santhosh Kumar K, Permaul K, Singh S (2011) High-level expression of a recombinant xylanase by Pichia pastoris NC38 in a 5L fermenter and its efficiency in biobleaching of bagasse pulp. Bioresour Technol 102(20):9723–9729
Collins T, Gerday C, Feller G (2005) Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev 29(1):3–23
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18(15):2714–2723
Harris GW, Pickersgill RW, Connerton I, Debeire P, Touzel JP, Breton C, Perez S (1997) Structural basis of the properties of an industrially relevant thermophilic xylanase. Proteins 29(1):77–86
Ko EP, Akatsuka H, Moriyama H, Shinmyo A, Hata Y, Katsube Y, Urabe I, Okada H (1992) Site-directed mutagenesis at aspartate and glutamate residues of xylanase from Bacillus pumilus. Biochem J 288(Pt 1):117–121
Liu YH, Lu FP, Li Y, Wang JL, Gao C (2008) Acid stabilization of Bacillus licheniformis alpha amylase through introduction of mutations. Appl Microbiol Biotechnol 80(5):795–803
Liu YH, Lu FP, Li Y, Yin XB, Wang Y, Gao C (2008) Characterisation of mutagenised acid-resistant alpha-amylase expressed in Bacillus subtilis WB600. Appl Microbiol Biotechnol 78(1):85–94
Matthews BW (1993) Structural and genetic analysis of protein stability. Annu Rev Biochem 62:139–160
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 13:426–428
Morris DD, Gibbs MD, Chin CW, Koh MH, Wong KK, Allison RW, Nelson PJ, Bergquist PL (1998) Cloning of the xynB gene from Dictyoglomus thermophilum Rt46B.1 and action of the gene product on kraft pulp. Appl Environ Microbiol 5(64):1759–1765
Morris DD, Gibbs MD, Ford M, Thomas J, Bergquist PL (1999) Family 10 and 11 xylanase genes from Caldicellulosiruptor sp. strain Rt69B.1. Extremophiles 3(2):103–111
Nakamura S, Nakai R, Namba K, Kubo T, Wakabayashi K, Aono R, Horikoshi K (1995) Structure-function relationship of the xylanase from alkaliphilic Bacillus sp. strain 41 M-1. Nucleic Acids Symp Ser 34:99–100
Nishimoto M, Kitaoka M, Hayashi K (2002) Employing chimeric xylanases to identify regions of an alkaline xylanase participating in enzyme activity at basic pH. J Biosci Bioeng 94(5):395–400
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31(13):3381–3385
Shao Q, Gao YQ (2011) The relative helix and hydrogen bond stability in the B domain of protein A as revealed by integrated tempering sampling molecular dynamics simulation. J Chem Phys 135(13):135102
Shirai T, Suzuki A, Yamane T, Ashida T, Kobayashi T, Hitomi J, Ito S (1997) High-resolution crystal structure of M-protease: phylogeny aided analysis of the high-alkaline adaptation mechanism. Protein Eng 10(6):627–634
Turunen O, Vuorio M, Fenel F, Leisola M (2002) Engineering of multiple arginines into the Ser/Thr surface of Trichoderma reesei endo-1,4-beta-xylanase II increases the thermotolerance and shifts the pH optimum towards alkaline pH. Protein Eng 15(2):141–145
Umemoto H, Ihsanawati Inami M, Yatsunami R, Fukui T, Kumasaka T, Tanaka N, Nakamura S (2009) Improvement of alkaliphily of Bacillus alkaline xylanase by introducing amino acid substitutions both on catalytic cleft and protein surface. Biosci Biotechnol Biochem 73(4):965–967
Wakarchuk W, Sung W, Campbell R, Cunningham A, Watson D, Yaguchi M (1994) Thermostabilization of the Bacillus circulans xylanase by the introduction of disulfide bonds. Protein Eng 7:1379–1386
Yang HM, Yao B, Meng K, Wang YR, Bai YG, Wu NF (2007) Introduction of a disulfide bridge enhances the thermostability of a Streptomyces olivaceoviridis xylanase mutant. J Ind Microbiol Biotechnol 34(3):213–218
Yazawa R, Takakura J, Sakata T, Ihsanawati, Yatsunami R, Fukui T, Kumasaka T, Tanaka N, Nakamura S (2011) A calcium-dependent xylan-binding domain of alkaline xylanase from alkaliphilic Bacillus sp. strain 41 M-1. Biosci Biotechnol Biochem 75(2):379–381
Zhang Z-G, Yi Z-L, Pei X-Q, Wu Z-L (2010) Improving the thermostability of Geobacillus stearothermophilus xylanase XT6 by directed evolution and site-directed mutagenesis. Bioresour Technol 101(23):9272–9278
Zheng H, liu Y, Liu X, Han Y, Wang J, Lu F (2012) Overexpression of a Paenibacillus campinasensis xylanase in Bacillus megaterium and its applications to biobleaching of cotton stalk pulp and saccharification of recycled paper sludge. Bioresour Technol 125:182–187
Zheng H, Liu Y, Liu X, Wang J, Han Y, Lu F (2012) Isolation, purification and characterization of a thermostable xylanase from a novel strain Paenibacillus campinasensis G1-1. J Microbiol Biotechnol 22(7):930–958
Acknowledgments
Financial support from the National High Technology Research and Development Program of China (Grant 2013AA102106), Program for Changjiang Scholars and Innovative Research Team in University (Grant IRT1166), the National Natural Science Fund (Grant 31,101,219) and China Postdoctoral Science Fund (Grant 2013M540076) are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
H. Zheng and Y. Liu, who contributed equally to this work, are both nominated as the first author.
Rights and permissions
About this article
Cite this article
Zheng, H., Liu, Y., Sun, M. et al. Improvement of alkali stability and thermostability of Paenibacillus campinasensis Family-11 xylanase by directed evolution and site-directed mutagenesis. J Ind Microbiol Biotechnol 41, 153–162 (2014). https://doi.org/10.1007/s10295-013-1363-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-013-1363-6