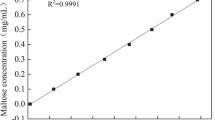
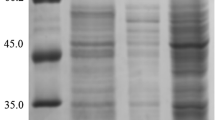
Abstract
Based on the original thermostable alpha-amylase gene from Bacillus licheniformis, two amino acids were site-directed mutagenised by polymerase chain reaction to obtain a new gene. This gene, with Leu134→Arg and Ser320→Ala, was substituted for acid-resistant capability previously. To favor purification of the product, high-level expression and secretion of mature, authentic and stable recombinant mutagenised alpha-amylase were achieved with protease-deficient strain Bacillus subtilis WB600 as the host. The recombinant mutagenised alpha-amylase with the activity of 4,700 U/mL was then purified by ammonium sulphate fractionation, anion exchange and gel filtration, consecutively. By multi-step purification, the specific activity of the recombinant protein was up to 916.7 U/mg with a 187.1-fold purification. The mutagenised protein was found to be more acid resistant than the native protein. The optimum pH and stable range of pH with the mutagenised protein was 4.5 and 4.0 to 6.5, respectively, compared with pH 6.5 and 5.5 to 7.0 as the favorite pH and pH stability range of the native protein.







Similar content being viewed by others
References
Arnold FH, Volkov AA (1999) Directed evolution of biocatalysts. Curr Opin Chem Biol 3:54–59
Bernstein FC, Koetzle TF, Williams GJ, Meyer Jr EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M (1977) The Protein Databank. A computer-based archival file for macromolecular structures. Eur J Biochem 80:319–324
Cai H, Chen ZJ, Du LX, Lu FP (2005) Expression and secretion of an acid-stable α-amylase gene in Bacillus subtilis by sacB promoter and signal peptide. Biotechnol Lett 27:1731–1736
Coco WM, Levinson WE, Crist MJ, Hektor HJ, Darzins A, Pienkos PT, Squires CH, Monticello DJ (2001) DNA shuffling method for generating highly recombined genes and evolved enzymes. Nat Biotechnol 19:354–359
Crabb WD, Shetty JK (1999) Commodity scale production of sugars from starches. Curr Opin Microbiol 2:252–256
Declerck N, Joyet P, Trosset JY, Garnier J, Gaillardin C (1995) Hyperthermostable mutants of Bacillus licheniformis α-amylase: multiple amino acid replacements and molecular modelling. Protein Eng 8:1029–1037
Declerck N, Machius M, Chambert R, Wiegand G, Huber R, Gaillardin C (1997) Hyperthermostable mutants of Bacillus licheniformis alpha-amylase: thermodynamic studies and structural interpretation. Protein Eng 10:541–549
Declerck N, Machius M, Wiegand G, Huber R, Gaillardin C (2000) Probing structural determinants specifying high thermostability in Bacillus licheniformis alpha-amylase. J Mol Biol 301:1041–1057
Fang TY, Ford C (1998) Protein engineering of Aspergillus awamori glucoamylase to increase its pH optimum. Protein Eng 11:383–388
Giver L, Gershenson A, Freskgard PO, Arnold FH (1998) Directed evolution of a thermostable esterase. Proc Natl Acad Sci USA 95:12809–12813
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Hartl B, Wehrl W, Wiegert T, Homuth G, Schumann W (2001) Development of a new integration site within the Bacillus subtilis chromosome and construction of compatible expression cassettes. J Bacteriol 183:2696–2699
Igarashi K, Hatada Y, Ikawa K, Araki H, Ozawa T, Kobayashi T, Ozaki K, Ito S (1998) Improved thermostability of an arginine-glycine residue is caused by enhanced calcium binding. Biochem Biophys Res Commun 248:372–377
Klein C, Hollender J, Bender H, Schulz GE (1992) Catalytic center of cyclodextrin glycosyltransferase derived from X-ray structure analysis combined with site-directed mutagenesis. Biochem 31:8740–8746
Knegtel RM, Strokopytov B, Penninga D, Faber OG, Rozeboom HJ, Kalk KH, Dijkhuizen L, Dijkstra BW (1995) Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products. J Biol Chem 270:29256–29264
Krengel U, Dijkstra BW (1996) Three-dimensional structure of endo-1,4-beta-xylanase I from Aspergillus niger: molecular basis for its low pH optimum. J Mol Biol 263:70–78
Kuchner O, Arnold FH (1997) Directed evolution of enzyme catalysts. Trends Biotechnol 15:523–530
Lee S, Oneda H, Minoda M, Tanaka A, Inouye K (2006) Comparison of starch hydrolysis activity and thermal stability of two Bacillus licheniformis α-amylases and insights into engineering-amylase variants active under acidic conditions. J Biol Chem 139:997–1005
Lowry OH, Rosbrough NL, Farr AL (1951) Protein measurement with folin phenol reagent. J Biol Chem 193:265–275
MacGregor EA (1993) Relationships between structure and activity in the α-amylase family of starch-metabolising enzymes. Starch 7:232–237
Machius M, Declerck N, Huber R, Wiegand G (2003) Kinetic stabilization of Bacillus licheniformis α-amylase through introduction of hydrophobic residues at the surface. J Biol Chem 278:11546–11553
Matsuzaki H, Yamane K, Yamaguchi K, Nagata Y, Maruo B (1974) Hybrid alpha-amylases produced by transformants of Bacillus subtilis. I. Purification and characterization of extracellular alpha-amylases produced by the parental strains and transformants. Biochim Biophys Acta 365:235–247
McCarter JD, Withers SG (1996) Unequivocal identification of Asp-214 as the catalytic nucleophile of Saccharomyces cerevisiae alphaglucosidase using 5-fluoroglycosyl fluorides. J Biol Chem 271:6889–6894
Meijer WJJ, Margarita S (2004) Relevance of UP elements for three strong Bacillus subtilis phage phi 29 promoters. Nucleic Acids Res 32:1166–1176
Nagano N, Porter CT, Thornton JM (2001) The (betaalpha) (8) glycosidases: sequence and structure analyses suggest distant evolutionary relationships. Protein Eng 14:845–855
Ness JE, Welch M, Giver L, Bueno M, Cherry JR, Borchert TV, Stemmer WP, Minshull J (1999) DNA shuffling of subgenomic sequences of subtilisin. Nat Biotechnol 17:893–896
Nielsen JE, Beier L, Otzen D, Borchert TV, Frantzen HB, Andersen KV, Svendsen A (1999) Electrostatics in the active site of an α-amylase. Eur J Biochem 264:816–824
Nielsen JE, Borchert TV (2000) Protein engineering of bacterial α-amylase. Biochim Biophys Acta 1543:253–274
Nielsen JE, Borchert TV, Vriend G (2001) The determinants of α-amylase pH-activity profiles. Protein Eng 14:505–512
Peitsch MC (1996) ProMod and Swiss-Model: internet-based tools for automated comparative protein modelling. Biochem Soc Trans 24:274–279
Peitsch MC, Wells TN, Stampf DR, Sussman JL (1995) The Swiss-3DImage collection and PDB-browser on the World-Wide Web. Trends Biochem Sci 20:82–84
Peitsch MC, Herzyk P, Wells TN, Hubbard RE (1996) Automated modelling of the transmembrane region of G-protein coupled receptor by Swiss-model. Receptors Channels 4:161–164
Qian M, Haser R, Buisson G, Duee E, Payan F (1994) The active center of a mammalian α-amylase. Structure of the complex of a pancreatic α-amylase with a carbohydrate inhibitor refined to 2.2A resolution. Biochem 33:6284–6294
Richardson TB, Tan XQ, Frey G, Callen W, Cabell M, Lam D, Macomber J, Short JM, Robertson DE, Miller C (2002) A novel, high performance enzyme for starch liquefaction. J Biol Chem 277:26501–26507
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory, New York
Saito N (1973) A thermophilic extracellular α-amylase from Bacillus licheniformis. Arch Biochem Biophys 155:290–298
Schmidt-Dannert C, Arnold FH (1999) Directed evolution of industrial enzymes. Trends Biotechnol 17:135–136
Shaw A, Bott R (1996) Engineering enzymes for stability. Curr Opin Biotechnol 6:546–550
Shaw A, Bott R, Day AG (1999) Protein engineering of α-amylase for low pH performance. Curr Opin Biotechnol 10:349–352
Strokopytov B, Penninga D, Rozeboom HJ, Kalk KH, Dijkhuizen L, Dijkstra BW (1995) X-ray structure of cyclodextrin glycosyltransferase complexed with acarbose. Implications for the catalytic mechanism of glycosidases. Biochem 34:2234–2240
Svensson B, Sogaard M (1992) Protein engineering of amylases. Biochem Soc Trans 20:34–42
Svensson B (1994) Protein engineering in the α-amylase family: catalytic mechanism, substrate specificity and stability. Plant Mol Biol 25:141–157
Uitdehaag JC, Mosi R, Kalk KH, van der Veen BA, Dijkhuizen L, Withers SG, Dijkstra BW (1999) X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat Struct Biol 6:432–436
Violet M, Meunier JC (1989) Kinetic study of the irreversible thermal denaturation of Bacillus licheniformis α-amylase. Biochem J 263:665–670
Wong SL, Kawamura F, Doi RH (1986) Use of the Bacillus subtilis subtilisin signal peptide for efficient secretion of TEMβ-lactamase during growth. J Bacteriol 168:1005–1009
Wong SL (1995) Advances in the use of Bacillus subtilis for the expression and secretion of heterologous proteins. Curr Opin Biotechnol 6:517–522
Wu SC, Wong SL (1999) Development of improved pUB110-based vectors for expression and secretion studies in Bacillus subtilis. J Biochem 72:185–195
Wu XC, Lee W, Tran L, Wong SL (1991) Engineering a Bacillus subtilis expression–secretion system with a strain deficient in six extracellular proteases. J Bacteriol 173:4952–4558
Yasbin RE, Wilson GA, Young FE (1975) Transformation and transfection in lysogenic strains of Bacillus subtilis: evidence for selective induction of prophage. J Bacteriol 121:296–304
Yuuki T, Nomura T, Tezuka H, Tsuboi A, Yamagata H, Tsukagoshi N, Udaka S (1985) Complete nucleotide sequence of a gene coding for heat-and pH-stable α-amylase of Bacillus licheniformis: comparison of the amino acid sequences of three bacterial liquefying α-amylase deduced from the DNA sequences. J Biochem 98:1147–1156
Zhang XZ, Cui ZL, Hong Q, Li SP (2005) High-level expression and secretion of methyl parathion hydrolase in Bacillus subtilis WB800. Appl Environ Microbiol 71:4101–4103
Acknowledgments
The authors would like to thank Dr. Sui-Lam Wong for the supply of plasmid pWB980. Dr. Sui-Lam Wong is a senior medical scholar from University of Calgary. This work was supported by Key Program of Tianjin Science and Technology Development Plan (06YFGPSH03500).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, Yh., Lu, Fp., Li, Y. et al. Characterisation of mutagenised acid-resistant alpha-amylase expressed in Bacillus subtilis WB600. Appl Microbiol Biotechnol 78, 85–94 (2008). https://doi.org/10.1007/s00253-007-1287-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-007-1287-z