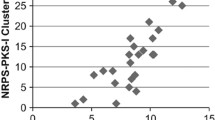
Abstract
Advances in DNA sequencing technologies have made it possible to sequence large numbers of microbial genomes rapidly and inexpensively. In recent years, genome sequencing initiatives have demonstrated that actinomycetes with large genomes generally have the genetic potential to produce many secondary metabolites, most of which remain cryptic. Since the numbers of new and novel pathways vary considerably among actinomycetes, and the correct assembly of secondary metabolite pathways containing type I polyketide synthase or nonribosomal peptide synthetase (NRPS) genes is costly and time consuming, it would be advantageous to have simple genetic predictors for the number and potential novelty of secondary metabolite pathways in targeted microorganisms. For secondary metabolite pathways that utilize NRPS mechanisms, the small chaperone-like proteins related to MbtH encoded by Mycobacterium tuberculosis offer unique probes or beacons to identify gifted microbes encoding large numbers of diverse NRPS pathways because of their unique function(s) and small size. The small size of the mbtH-homolog genes makes surveying large numbers of genomes straight-forward with less than ten-fold sequencing coverage. Multiple MbtH orthologs and paralogs have been coupled to generate a 24-mer multiprobe to assign numerical codes to individual MbtH homologs by BLASTp analysis. This multiprobe can be used to identify gifted microbes encoding new and novel secondary metabolites for further focused exploration by extensive DNA sequencing, pathway assembly and annotation, and expression studies in homologous or heterologous hosts.



Similar content being viewed by others
References
Albright JC, Goering AW, Doroghazi JR, Metcalf WW, Kelleher NL (2013) Strain-specific proteogenomics accelerates discovery of natural products via their biosynthetic pathways. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1373-4
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Baltz RH (2005) Antibiotic discovery from actinomycetes: will a renaissance follow the decline and fall? SIM News 55:186–196
Baltz RH (2007) Antimicrobials from actinomycetes: back to the future. Microbe 2:125–131
Baltz RH (2008) Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol 8:557–563
Baltz RH (2010) Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol 37:759–772
Baltz RH (2010) Genomics and the ancient origins of the daptomycin biosynthetic gene cluster. J Antibiot 63:506–511
Baltz RH (2011) Strain improvement in actinomycetes in the postgenomic era. J Ind Microbiol Biotechnol 38:657–666
Baltz RH (2011) Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. J Ind Microbiol Biotechnol 38:1747–1760
Bentley SD, Chater KF, Cerdeño-Tárraga AM et al (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Blodgett JA, Zhang JK, Metcalf WW (2005) Molecular cloning, sequence analysis, and heterologous expression of the phosphinothricin tripeptide biosynthetic gene cluster from Streptomyces viridochromogenes DSM 40736. Antimicrob Agents Chemother 49:230–240
Blodgett JAV, Oh D-C, Cao S, Currie CR, Kolter R, Clardy J (2010) Common biosynthetic origins for polycyclic tetramate macrolactams from phylogenetically diverse bacteria. Proc Natl Acad Sci USA 107:11692–11697
Boddy CN (2013) Bioinformatics tools for genome mining of polyketide and non-ribosomal peptides. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1368-1
Boll B, Heide L (2013) A domain of RubC1 biosynthesis that can functionally replace MbtH-like proteins in tyrosine adenylation. ChemBioChem 14:43–44
Challis GL (2008) Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology 154:1555–1569
Chen H, Hubbard BK, O’Connor SE, Walsh CT (2002) Formation of β-hydroxy histidine in the biosynthesis of nikkomycin antibiotics. Chem Biol 9:103–112
Choo SW, Wong YL, Yusoff AM, Leong ML, Wong GJ, Ong CS, Ng KP, Ngeow YF (2012) Genome sequence of the Mycobacterium abscessus strain 93. J Bacteriol 194:3278
Corre C, Challis GL (2009) New natural product biosynthetic chemistry discovered by genome mining. Nat Prod Rep 26:977–986
Davidsen JM, Bartley DM, Townsend CA (2013) Non-ribosomal propeptide precursor of nocardicin A biosynthesis predicted from adenylation domain specificity dependent on the MbtH family protein NocI. J Am Chem Soc 135:1749–1759
Demain AL (2013) Importance of microbial natural products and the need to revitalize their discovery. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1325-z
Donadio S, Monciardini P, Sosio M (2007) Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat Prod Rep 24:1073–1109
Du L, Sánchez C, Chen M, Edwards DJ, Shen B (2000) The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem Biol 7:623–642
Felnagle EA, Rondon MR, Berti AD, Crosby HA, Thomas MG (2007) Identification of the biosynthetic gene cluster and an additional gene for resistance to the antituberculosis drug capreomycin. Appl Environ Microbiol 73:4162–4170
Felnagle EA, Podevels AM, Barkei JJ, Thomas MG (2011) Mechanistically distinct nonribosomal peptide synthetases assemble the structurally related viomycin and capreomycin antibiotics. ChemBioChem 12:1859–1867
Galm U, Schimana J, Fiedler H-P, Schmidt J, Li S-M, Heide L (2002) Cloning and analysis of the simocyclinone biosynthetic gene cluster of Streptomyces antibioticus Tu 6040. Arch Microbiol 178:102–114
Galm U, Wendt-Pienkowski E, Wang L, Oh T-J, Yi F, Tao M, Coughlin JM, Shen B (2009) The biosynthetic gene cluster of zorbamycin, a member of the bleomycin family of antitumor antibiotics, from Streptomyces flavoviridis ATTC 21892. Mol BioSyst 5:77–90
Galm U, Wendt-Pienkowski E, Wang L, Huang S-X, Unsin C, Tao M, Coughlin JM, Shen B (2011) Comparative analysis of the biosynthetic gene clusters and pathways for three structurally related antitumor antibiotics: bleomycin, tallysomycin, and zorbamycin. J Nat Prod 74:526–536
Giddings LA, Newman DJ (2013) Microbial natural products: molecular blueprints for antitumor drugs. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1331-1
Gomez-Escribano JP, Bibb MJ (2013) Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: from genome mining to manipulation of biosynthetic pathways. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1348-5
Herbst DA, Boll B, Zocher G, Stehle T, Heide L (2013) Structural basis of the interaction of MbtH-like proteins, putative regulators of nonribosomal peptide biosynthesis, with adenylating enzymes. J Biol Chem 288:1991–2003
Higgs RE, Zahn JA, Gygi FD, Hilton MD (2001) Rapid method to estimate the presence of secondary metabolites in microbial extracts. Appl Environ Microbiol 67:371–376
Hojati Z, Milne C, Harvey B, Gordon L, Borg M, Flett F, Wilkinson B, Sidebottom PJ, Rudd BA, Hayes MA, Smith CP, Micklefield J (2002) Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem Biol 9:1175–1187
Ikeda H, Shinya K, Ōmura S (2013) Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1327-x
Kaysser L, Wemakor E, Sedding K, Hennig S, Siedenberg S, Gust B (2011) Identification of a napsamycin biosynthetic gene cluster by genome mining. ChemBioChem 12:477–478
Keller U, Lang M, Crnovcic I, Pfennig F, Schauwecker F (2010) The actinomycin biosynthetic gene cluster of Streptomyces chrysomallus: a genetic hall of mirrors for the synthesis of a molecule with mirror symmetry. J Bacteriol 192:2583–2595
Land M, Lapidus A, Mayilraj S et al (2009) Complete genome sequence of Actinosynnema mirum type strain (101). Stand Genomic Sci 1:46–53
Lauer B, Russwurm R, Bormann C (2000) Molecular characterization of two genes from Streptomyces tendae Tü901 required for the formation of the 4-formyl-4-imidazolin-2-one-containing nucleoside moiety of the peptidyl nucleoside antibiotic nikkomycin. Eur J Biochem 267:1698–1706
Lauer B, Russwurm R, Schwarz W, Kálmánczhelyi A, Bruntner C, Rosemeier A, Bormann C (2001) Molecular characterization of co-transcribed genes from Streptomyces tendae Tü901 involved in the biosynthesis of the peptidyl moiety and assembly of the peptidyl nucleoside antibiotic nikkomycin. Mol Gen Genet 264:662–673
Lautru S, Oves-Costales D, Pernodet J-L, Challis GL (2007) MbtH-like protein-mediated cross-talk between non-ribosomal peptide antibiotic and siderophore biosynthetic pathways in Streptomyces coelicolor M145. Microbiology 153:1405–1412
Li L, Deng W, Song J, Ding W, Zhao Q-F, Peng C, Song W-W, Tang T-L, Liu W (2008) Characterization of the saframycin A gene cluster from Streptomyces lavendulae NRRL 110002 revealing a nonribosomal peptide synthetase system for assembling the unusual tetrapeptidyl skeleton in an iterative manner. J Bacteriol 190:251–263
Liu WT, Kerten RD, Yang YL, Moore BS, Dorrestein PC (2011) Imaging mass spectrometry and genome mining via short sequence tagging identified the anti-infective agent arylomycin in Streptomyces roseosporus. J Am Chem Soc 133:18010–18013
Lombó F, Velasco A, Castro A, de la Calle F, Braña AF, Sánchez-Puelles JM, Méndez C, Salas JA (2006) Deciphering the biosynthesis pathway of the antitumor thiocoraline from a marine actinomycete and its expression in two Streptomyces species. ChemBioChem 7:366–376
Luzhetskyy A, Rebets Y, Brötz E, Tokovenko B (2013) Actinomycetes biosynthetic potential: how to bridge in silico and in vivo. J Ind Microbiol Biotechnol. doi:10-1007/s10295-013-1352-9
Magarvey NA, Haltli B, He M, Greenstein M, Hucul JA (2006) Biosynthetic pathway for mannopeptimycins, lipoglycopeptide antibiotics active against drug-resistant Gram-positive pathogens. Antimicrob Agents Chemother 50:2167–2177
Mast Y, Weber T, Golz M, Ort-Winklbauer R, Gondran A, Wohlleben W, Schinko E (2011) Characterization of the ‘pristinamycin supercluster’ of Streptomyces pristinaespiralis. Microb Biotechnol 4:192–206
Mast YJ, Wohlleben W, Schinko E (2011) Identification and functional characterization of phenylglycine biosynthetic genes involved in pristinamycin biosynthesis in Streptomyces pristinaespiralis. J Biotechnol 155:63–67
Miao V, Coëffet-LeGal M-F, Brian P, Brost R, Penn J, Whiting A, Martin S, Ford R, Parr I, Bouchard M, Silva CJ, Wrigley SK, Baltz RH (2005) Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 151:1507–1523
Miao V, Brost R, Chapple J, She K, Coëffet-Le Gal M-F, Baltz RH (2006) The lipopeptide antibiotic A54145 biosynthetic gene cluster from Streptomyces fradiae. J Ind Microbiol Biotechnol 33:129–140
Müller C, Nolden S, Gebhardt P, Heinzelmann E, Lange C, Puk O, Welzel K, Wohlleben W, Schwartz D (2007) Sequencing and analysis of the biosynthetic gene cluster of the lipopeptide antibiotic friulimicin in Actinoplanes friuliensis. Antimicrob Agents Chemother 51:1028–1037
Nett M, Ikeda H, Moore BS (2009) Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26:1362–1384
Newman DJ, Cragg GM (2012) Natural products as sources of new drugs over the 30 years from 1981–2010. J Nat Prod 75:311–335
Ningsih F, Kitani S, Fukushima E, Nihira T (2011) VisG is essential for biosynthesis of virginiamycin S, a streptogramin type B antibiotic, as a provider of the nonproteogenic amino acid phenylglycine. Microbiology 157:3213–3220
Nolan M, Sikorski J, Janko M et al (2010) Complete genome sequence of Streptosporangium roseum type strain (NI 9100). Stand Genome Sci 2:29–37
Ochi K, Tanaka Y, Tojo S (2013) Activating the expression of cryptic genes by rpoB mutations in RNA polymerase or by rare earth elements. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1349-4
Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S (2008) Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol 190:4050–4060
Olano C, Gómez C, Pérez M, Palomino M, Pineda-Lucena A, Carbajo RJ, Braña AF, Méndez C, Salas JA (2009) Deciphering the biosynthesis of the RNA polymerase inhibitor streptolydigin and generation of glycosylated derivatives. Chem Biol 16:1031–1044
Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF (2007) Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol 25:447–453
Patzer SI, Braun V (2010) Gene cluster involved in the biosynthesis of griseobactin, a catechol-peptide siderophore of Streptomyces sp. ATCC 700974. J Bacteriol 192:426–435
Pojer F, Li S-M, Heide L (2002) Molecular cloning and sequence analysis of the clorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. Microbiology 148:3901–3911
Praseuth AP, Wang CC, Watanabe K, Hotta K, Oguri H, Oikawa H (2008) Complete sequence of biosynthetic gene cluster responsible for producing triostin A and evaluation of quinomycin-type antibiotics from Streptomyces trinostinicus. Biotechnol Prog 24:1226–1231
Qin S, Zhang H, Li F, Zhu B, Zheng H (2012) Draft genome sequence of marine Streptomyces sp. Strain W007, which produces angucyclinone antibiotics with a benz[a]anthracene skeleton. J Bacteriol 194:1628–1629
Qu X, Jiang N, Xu F, Shao L, Tang G, Wilkensen B, Liu W (2011) Cloning, sequencing and characterization of the biosynthetic gene cluster of sanglifehrin A, a potent cyclophilin inhibitor. Mol BioSyst 7:852–861
Quadri LE, Sello J, Keating TA, Weinreb PH, Walsh CT (1998) Identification of Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem Biol 5:631–645
Rackham EJ, Gruschow S, Ragab AE, Dickens S, Goss RJM (2010) Pacidamycin biosynthesis: identification and heterologous expression of the first uridyl peptide antibiotic gene cluster. ChemBioChem 11:1700–1709
Schweintek P, Szczepanowski R, Ruckert C et al (2012) The complete genome sequence of the acarbose producer Actinoplanes sp. SE50/110. BMC Genomics 13:112
Sosio M, Stinchi S, Beltrametti F, Lazzarini A, Donadio S (2003) The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by Nonomuraea species. Chem Biol 10:541–549
Sosio M, Kloosterman H, Bianchi A, de Vreugd P, Dijkhuizen L, Donodio S (2004) Organization of the teicoplanin gene cluster in Actinoplanes teichomyceticus. Microbiology 150:95–102
Stegmann E, Rausch C, Stockert S, Burkert D, Wohlleben W (2006) The small MbtH-like protein encoded by an internal gene of the balhimycin biosynthetic gene cluster is not required for glycopeptide production. FEMS Microbiol Lett 262:85–92
Strobel T, Al-Dilaimi A, Blom J, Gessner A, Kalinowski J, Luzhetska M, Pühler A, Szczepanowski R, Bechtold A, Rücker C (2012) Complete genome sequence of Saccharothrix espanaensis DSM 44229 and comparison to the other completely sequenced Pseudonocardiaceae. BMC Genomics 13:465
Tao M, Wang L, Wendt-Pienkowski E, George NP, Galm U, Zhang G, Coughlin JM, Shen B (2007) The tallysomycin biosynthetic gene cluster from Streptoalloteichus hindustanus E465-94 ATTC 31158 unveiling new insights into the biosynthesis of the bleomycin family of antitumor antibiotics. Mol BioSyst 3:60–74
Thomas MG, Chan YA, Ozanick SG (2003) Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster. Antimicrob Agents Chemother 47:2823–2830
Wang G, Nie L, Tan H (2003) Cloning and characterization of sanO, a gene involved in nikkomycin biosynthesis in Streptomyces ansochromogenes. Lett Appl Microbiol 37:452–457
Wang L, Wang S, He Q, Yu T, Li Q, Hong B (2012) Draft genome sequence of Streptomyces globisporus C-1027, which produces an antitumor antibiotic consisting of a nine-membered enediyne with a chromoprotein. J Bacteriol 194:4144
Wang L, Xie Y, Li Q, He N, Yao E, Xu H, Yu Y, Chen R, Hong B (2012) Draft genome sequence of Streptomyces sp. Strain SS, which produces a series of uridyl peptide antibiotic sansanmycins. J Bacteriol 194:6988–6989
Wang Y, Chen Y, Shen Q, Yin X (2011) Molecular cloning and identification of the laspartomycin biosynthetic gene cluster from Streptomyces viridochromogenes. Gene 483:11–21
Wang Z-X, Li S-M, Heide L (2000) Identification of the coumermycin A1 biosynthetic gene cluster of Streptomyces rishiriensis DSM 40489. Antimicrob Agents Chemother 44:3040–3048
Weber T, Laiple KJ, Pross EK, Textor A, Grond S, Welzel K, Pelzer S, Vente A, Wohlleben W (2008) Molecular analysis of the kirromycin biosynthetic gene cluster revealed beta-alanine as precursor of the pyridone moiety. Chem Biol 15:175–188
Yang H, He T, Zhu W, Lu B, Sun W (2013) Whole-genome shotgun assembly and analysis of the genome of Streptomyces mobaraensis DSM 40847, a strain for the industrial production of microbial transglutaminase. Genome Announc 1:e0014313
Yarbrough GG, Taylor DP, Rowlands RT, Crawford MS, Lasure LL (1992) Screening microbial metabolites for new drugs—theoretical and practical issues. J Antibiot 46:535–544
Yin X, Zabriskie M (2006) The enduracidin biosynthetic gene cluster from Streptomyces fungicidicus. Microbiology 152:2969–2983
Zahn JA, Higgs RE, Hilton MD (2001) Use of direct-infusion electrospray mass spectrometry to guide empirical development of improved conditions for expression of secondary metabolites from actinomycetes. Appl Environ Microbiol 67:377–386
Zerbe K, Pylypenko O, Vitali F, Zhang W, Rouset S, Heck M, Vrijbloed JW, Biscoff D, Bister B, Sussmuth RD, Pelzer S, Wohlleben W, Robinson JA, Schlichting I (2002) Crystal structure of OxyB, a cytochrome P450 implicated in an oxidative phenol coupling reaction during vancomycin biosynthesis. J Biol Chem 277:47476–47485
Zerikly M, Challis GL (2009) Strategies for the discovery of new natural products by genome mining. ChemBioChem 10:625–633
Zhang C, Kong L, Liu Q, Lei X, Zhu T, Yin J, Lin B, Deng Z, You D (2013) In vitro characterization of echinomycin biosynthesis: formation and hydroxylation of l-tryptophanyl-S-enzyme and oxidation of (2S,3S) β-hydroxytryptophan. PLOS ONE 8:e56772
Zhang W, Ostach B, Walsh CT (2010) Identification of the biosynthetic gene cluster for the pacidamycin group of peptidyl nucleoside antibiotics. Proc Natl Acad Sci USA 107:16828–16833
Zhang W, Heemstra JR, Walsh CT, Imker HJ (2010) Activation of the pacidamycin PacL adenylation domain by MbtH-like proteins. Biochemistry 49:9946–9947
Zhao W, Zhong Y, Yuan H et al (2010) Complete genome sequence of the rifamycin SV-producing Amycolatopsis mediterranei U32 revealed its genetic characteristics in phylogeny and metabolism. Cell Res 20:1096–1108
Zhu F, Qin C, Tao L et al (2011) Clustered patterns of species origins of nature-derived drugs and clues for future bioprospecting. Proc Natl Acad Sci USA 31:12943–12948
Zhu H, Sandiford SK, van Wezel GP (2013) Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol. doi:10.1007/s10295-013-1309-z
Acknowledgments
This work was supported by CognoGen Biotechnology Consulting.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Baltz, R.H. MbtH homology codes to identify gifted microbes for genome mining. J Ind Microbiol Biotechnol 41, 357–369 (2014). https://doi.org/10.1007/s10295-013-1360-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-013-1360-9