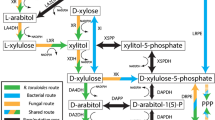
Abstract
In this report, a novel zymogram assay and coupled phosphoketolase assay were employed to demonstrate that Clostridium acetobutylicum gene CAC1343 encodes a bi-functional xylulose-5-P/fructose-6-P phosphoketolase (XFP). The specific activity of purified recombinant XFP was 6.9 U/mg on xylulose-5-P and 21 U/mg on fructose-6-P, while the specific activity of XFP in concentrated C. acetobutylicum whole-cell extract was 0.094 and 0.52 U/mg, respectively. Analysis of crude cell extracts indicated that XFP activity was present in cells grown on arabinose but not glucose and quantitative PCR was used to show that CAC1343 mRNA expression was induced 185-fold during growth on arabinose when compared to growth on glucose. HPLC analysis of metabolites revealed that during growth on xylose and glucose more butyrate than acetate was formed with final acetate:butyrate ratios of 0.72 and 0.83, respectively. Growth on arabinose caused a metabolic shift to more oxidized products with a final acetate:butyrate ratio of 1.95. The shift towards more oxidized products is consistent with the presence of an XFP, suggesting that arabinose is metabolized via a phosphoketolase pathway while xylose is probably metabolized via the pentose phosphate pathway.



Similar content being viewed by others
References
Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A (2011) Plant cell walls. Garland Science, Taylor and Francis Group, LLC, New York, NY
Edgar R, Domrachev M, Lash AE (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30(1):207–210
Finch AS, Mackie TD, Sund CJ, Sumner JJ (2011) Metabolite analysis of Clostridium acetobutylicum: fermentation in a microbial fuel cell. Bioresource Technol 102(1):312–315. doi:10.1016/j.biortech.2010.06.149
Gheshlaghi R, Scharer JM, Moo-Young M, Chou CP (2009) Metabolic pathways of clostridia for producing butanol. Biotechnol Adv 27(6):764–781. doi:10.1016/j.biotechadv.2009.06.002
Goldberg ML, Racker E (1962) Formation and isolation of a glycolaldehyde-phosphoketolase intermediate. J Biol Chem 237:3841–3842
Grimmler C, Held C, Liebl W, Ehrenreich A (2010) Transcriptional analysis of catabolite repression in Clostridium acetobutylicum growing on mixtures of d-glucose and d-xylose. J Biotechnol 150(3):315–323. doi:10.1016/j.jbiotec.2010.09.938
Gu Y, Li J, Zhang L, Chen J, Niu L, Yang Y, Yang S, Jiang W (2009) Improvement of xylose utilization in Clostridium acetobutylicum via expression of the talA gene encoding transaldolase from Escherichia coli. J Biotechnol 143(4):284–287. doi:10.1016/j.jbiotec.2009.08.009
Jones DT, Woods DR (1986) Acetone-butanol fermentation revisited. Microbiol Rev 50(4):484–524
Jurgens G, Survase S, Berezina O, Sklavounos E, Linnekoski J, Kurkijarvi A, Vakeva M, van Heiningen A, Granstrom T (2012) Butanol production from lignocellulosics. Biotechnol Lett 34(8):1415–1434. doi:10.1007/s10529-012-0926-3
Kim BH, Gadd GM (2008) Bacterial physiology and metabolism. Cambridge University Press, Cambridge
Lee J, Yun H, Feist AM, Palsson BO, Lee SY (2008) Genome-scale reconstruction and in silico analysis of the Clostridium acetobutylicum ATCC 824 metabolic network. Appl Microbiol Biot 80(5):849–862. doi:10.1007/s00253-008-1654-4
Lee JY, Jang YS, Lee J, Papoutsakis ET, Lee SY (2009) Metabolic engineering of Clostridium acetobutylicum M5 for highly selective butanol production. Biotechnol J 4(10):1432–1440. doi:10.1002/biot.200900142
Lutke-Eversloh T, Bahl H (2011) Metabolic engineering of Clostridium acetobutylicum: recent advances to improve butanol production. Curr Opin Biotechnol 22(5):634–647. doi:10.1016/j.copbio.2011.01.011
Manchenko GP (2003) Handbook of detection of enzymes on electrophoretic gels, 2nd edn. CRC Press, Boca Raton
Meile L, Rohr LM, Geissman TA, Herensperger M, Teuber M (2001) Characterization of the d-xylulose 5-phosphate/d-fructose 6-phosphate phosphoketolase gene (xfp) from Bifidobacterium lactis. J Bacteriol 183(9):2929–2936
Nigam PS, Singh A (2011) Production of liquid biofuels from renewable resources. Prog Energ Combust 37(1):52–68. doi:10.1016/j.pecs.2010.01.003
Ounine K, Petitdemange H, Raval G, Gay R (1983) Acetone-butanol production from pentoses by Clostridium Acetobutylicum. Biotechnol Lett 5(9):605–610
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45
Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339(1):62–66
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Sanchez B, Zuniga M, Gonzalez-Candelas F, de los Reyes-Gavilan CG, Margolles A (2010) Bacterial and eukaryotic phosphoketolases: phylogeny, distribution and evolution. J Mol Microb Biotech 18(1):37–51. doi:10.1159/000274310
Senger RS, Papoutsakis ET (2008) Genome-scale model for Clostridium acetobutylicum: part I. Metabolic network resolution and analysis. Biotechnol Bioeng 101 (5):1036–1052. doi:10.1002/Bit.22010
Servinsky MD, Kiel JT, Dupuy NF, Sund CJ (2010) Transcriptional analysis of differential carbohydrate utilization by Clostridium acetobutylicum. Microbiol-Sgm 156:3478–3491. doi:10.1099/Mic.0.037085-0
Shinkawa S, Okano K, Yoshida S, Tanaka T, Ogino C, Fukuda H, Kondo A (2011) Improved homo l-lactic acid fermentation from xylose by abolishment of the phosphoketolase pathway and enhancement of the pentose phosphate pathway in genetically modified xylose-assimilating Lactococcus lactis. Appl Microbiol Biot 91(6):1537–1544. doi:10.1007/s00253-011-3342-z
Wiesenborn DP, Rudolph FB, Papoutsakis ET (1988) Thiolase from Clostridium-acetobutylicum Atcc-824 and its role in the synthesis of acids and solvents. Appl Environ Microbiol 54(11):2717–2722
Xiao H, Gu Y, Ning Y, Yang Y, Mitchell WJ, Jiang W, Yang S (2011) Confirmation and elimination of xylose metabolism bottlenecks in glucose phosphoenolpyruvate-dependent phosphotransferase system-deficient Clostridium acetobutylicum for simultaneous utilization of glucose, xylose, and arabinose. Appl Environ Microbiol 77(22):7886–7895. doi:10.1128/AEM.00644-11
Zhang L, Leyn SA, Gu Y, Jiang W, Rodionov DA, Yang C (2012) Ribulokinase and transcriptional regulation of arabinose metabolism in Clostridium acetobutylicum. J Bacteriol 194(5):1055–1064. doi:10.1128/JB.06241-11
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Servinsky, M.D., Germane, K.L., Liu, S. et al. Arabinose is metabolized via a phosphoketolase pathway in Clostridium acetobutylicum ATCC 824. J Ind Microbiol Biotechnol 39, 1859–1867 (2012). https://doi.org/10.1007/s10295-012-1186-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-012-1186-x