Abstract
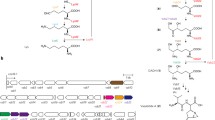
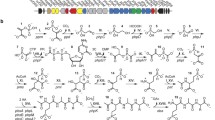
Polyketide antibiotics are among the most important therapeutics used in human and animal health care. Type II polyketides are composed primarily of acetate-derived thioesters, and the subunits for the PKS are contained in a single module that includes a ketosynthase, acyl carrier protein, chain-length factor and sometimes a keto-reductase, aromatase, cyclase and modifying enzymes, such as glycosylases or hydroxylases. While the enzyme complexes that make up the PKS have been the focus of intense study (Khosla in Chem Rev 7:2577–2590, 1997), the pathways for precursor synthesis have not been established and predictions are complicated by the fact that acetate may be derived from a number of metabolic pathways. Here we show that 50% of the acetate for synthesis of the Type II polyketide, actinorhodin, in Streptomyces coelicolor, is derived from the catabolism of the branched amino acids by pathways that are nutrient dependent. The streptomycetes are apparently unique in that they contain two BCDH gene clusters, each of which is potentially capable of converting leucine, valine and isoleucine to the corresponding thioesters, and contain at least three different pathways for valine catabolism that are differentially used in response to nutrient availability.





Similar content being viewed by others
References
Brana AF, Manzanal MB, Hardisson C (1982) Characterization of intracellular polysaccharides of Streptomyces. Can J Microbiol 28:1320–1323
Brawner ME, Auerbach JI, Fornwald JA, Rosenberg M, Taylor DP (1985) Characterization of Streptomyces promoter sequences using the Escherichia coli galactokinase gene. Gene 40:191–201. doi:10.1016/0378-1119(85)90042-3
Burke J, Schneider D, Westpheling J (2001) Generalized transduction in Streptomyces coelicolor. Proc Natl Acad Sci USA 98:6289–6294. doi:10.1073/pnas.101589398
Carreras CW, Khosla C (1998) Purification and in vitro reconstitution of the essential protein components of an aromatic polyketide synthase. Biochemistry 37:2084–2088. doi:10.1021/bi972919+
Cropp TA, Smogowicz AA, Hafner EW, Denoya CD, McArthur HA, Reynolds KA (2000) Fatty-acid biosynthesis in a branched-chain α-keto acid dehydrogenase mutant of Streptomyces avermitilis. Can J Microbiol 46:506–514. doi:10.1139/cjm-46-6-506
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645. doi:10.1073/pnas.120163297
Denoya CD, Fedechko RW, Hafner EW, McArthur HA, Morgenstern MR, Skinner DD, Stutzman-Engwall K, Wax RG, Wernau WC (1995) A second branched-chain alpha-keto acid dehydrogenase gene cluster (bkdFGH) from Streptomyces avermitilis: its relationship to avermectin biosynthesis and the construction of a bkdF mutant suitable for the production of novel antiparasitic avermectins. J Bacteriol 177:3504–3511
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA 100:1541–1546. doi:10.1073/pnas.0337542100
Hey-Ferguson A, Mitchell M, Elbein AD (1973) Trehalose metabolism in germinating spores of Streptomyces hygroscopicus. J Bacteriol 116:1084–1085
Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM, Schrempf H (1985) genetic manipulation of Streptomyces: a laboratory manual, 1st edn. The John Innes Foundation, Norwich
Kaneda T (1991) Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol Rev 55:288–302
Khosla C (1997) Harnessing the biosynthetic potential of modular polyketide synthases. Chem Rev 7:2577–2590. doi:10.1021/cr960027u
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces Genetics. The John Innes Foundation, Norwich
Li C, Akopiants K, Reynolds KA (2006) Identification and disruptional analysis of the Streptomyces cinnamonensis msdA gene, encoding methylmalonic acid semialdehyde dehydrogenase. J Ind Microbiol Biotechnol 33:75–83. doi:10.1007/s10295-005-0053-4
MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T (1992) Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61–68. doi:10.1016/0378-1119(92)90603-M
Moore BS, Hertweck C (2002) Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat Prod Rep 19:70–99. doi:10.1039/b003939j
Ratnatilleke A, Vrijbloed JW, Robinson JA (1999) Cloning and sequencing of the coenzyme B12-binding domain of isobutyryl-CoA mutase from Streptomyces cinnamonensis, reconstitution of mutase activity, and characterization of the recombinant enzyme produced in Escherichia coli. J Biol Chem 274:31679–31685. doi:10.1074/jbc.274.44.31679
Sambrook J, Russell D (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sherman MM, Yue S, Hutchinson CR (1986) Biosynthesis of lasalocid A. Metabolic interrelationships of carboxylic acid precursors and polyether antibiotics. J Antibiot (Tokyo) 39:1135–1143
Skinner DD, Morgenstern MR, Fedechko RW, Denoya CD (1995) Cloning and sequencing of a cluster of genes encoding branched-chain alpha-keto acid dehydrogenase from Streptomyces avermitilis and the production of a functional E1 [αβ] component in Escherichia coli. J Bacteriol 177:183–190
Sprusansky O, Stirrett K, Skinner D, Denoya C, Westpheling J (2005) The bkdR gene of Streptomyces coelicolor is required for morphogenesis and antibiotic production and encodes a transcriptional regulator of a branched-chain amino acid dehydrogenase complex. J Bacteriol 187:664–671. doi:10.1128/JB.187.2.664-671.2005
Strauch E, Takano E, Baylis HA, Bibb MJ (1991) The stringent response in Streptomyces coelicolor A3(2). Mol Microbiol 5:289–298. doi:10.1111/j.1365-2958.1991.tb02109.x
Tang L, Zhang YX, Hutchinson CR (1994) Amino acid catabolism and antibiotic synthesis: valine is a source of precursors for macrolide biosynthesis in Streptomyces ambofaciens and Streptomyces fradiae. J Bacteriol 176:6107–6119
Vrijbloed JW, Zerbe-Burkhardt K, Ratnatilleke A, Grubelnik-Leiser A, Robinson JA (1999) Insertional inactivation of methylmalonyl coenzyme A (CoA) mutase and isobutyryl-CoA mutase genes in Streptomyces cinnamonensis: Influence of polyketide antibiotic biosynthesis. J Bacteriol 181:5600–5605
Wallace KK, Zhao B, McArthur HA, Reynolds KA (1995) In vivo analysis of straight-chain and branched-chain fatty acid biosynthesis in three actinomycetes. FEMS Microbiol Lett 131:227–234. doi:10.1111/j.1574-6968.1995.tb07781.x
Willecke K, Pardee AB (1971) Fatty acid-requiring mutant of Bacillus subtilis defective in branched-chain α-keto acid dehydrogenase. J Biol Chem 246:5264–5272
Zhang YX, Tang L, Hutchinson CR (1996) Cloning and characterization of a gene (msdA) encoding methylmalonic acid semialdehyde dehydrogenase from Streptomyces coelicolor. J Bacteriol 178:490–495
Acknowledgments
We thank Kevin Reynolds and Galina Florova for performing the fatty acid analysis and for assistance in interpreting the data, Timothy Dore for his help in interpretation of the fatty acid analysis, Larry Shimketts and Mike Adams for advice throughout the course of the work and for critical review of the manuscript, and David Brown for help in preparation of the figures. K. S. was supported by a predoctoral training grant from the National Institute for General Medical Sciences GM07103 to the Genetics Department of the University of Georgia, an ARCS Foundation Scholarship, as well as a grant from Pfizer, Inc. to J. W.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Stirrett, K., Denoya, C. & Westpheling, J. Branched-chain amino acid catabolism provides precursors for the Type II polyketide antibiotic, actinorhodin, via pathways that are nutrient dependent. J Ind Microbiol Biotechnol 36, 129–137 (2009). https://doi.org/10.1007/s10295-008-0480-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-008-0480-0