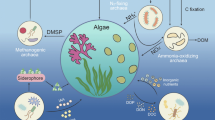
Abstract
This study reviews the available molecular methods and new high-throughput technologies for their practical use in the molecular detection, quantification, and diversity assessment of microalgae. Molecular methods applied to other groups of organisms can be adopted for microalgal studies because they generally detect universal biomolecules, such as nucleic acids or proteins. These methods are primarily related to species detection and discrimination among various microalgae. Among current molecular methods, some molecular tools are highly valuable for small-scale detection [e.g., single-cell polymerase chain reaction (PCR), quantitative real-time PCR (qPCR), and biosensors], whereas others are more useful for large-scale, high-throughput detection [e.g., terminal restriction length polymorphism, isothermal nucleic acid sequence-based amplification, loop-mediated isothermal amplification, microarray, and next generation sequencing (NGS) techniques]. Each molecular technique has its own strengths in detecting microalgae, but they may sometimes have limitations in terms of detection of other organisms. Among current technologies, qPCR may be considered the best method for molecular quantification of microalgae. Metagenomic microalgal diversity can easily be achieved by 454 pyrosequencing rather than by the clone library method. Current NGS, third and fourth generation technologies pave the way for the high-throughput detection and quantification of microalgal diversity, and have significant potential for future use in field monitoring.

Similar content being viewed by others
References
Ahn S, Kulis DM, Erdner DL, Anderson DM, Walt DR (2006) Fiber-optic for the simultaneous detection of multiple harmful algal bloom species. Appl Environ Microb 72(9):5742–5749
Alpermann TJ, Tillmann U, Beszteri B, Cembella AD, John U (2010) Phenotypic variation and genotypic diversity in a planktonic population of the toxigenic marine dinoflagellate Alexandrium tamarense (Dinophyceae). J Phycol 46:18–32
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59(1):143–169
Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM (2009) A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One 4(7):e6372
Anderson DM (1995) Identification of harmful algal species using molecular probes: an emerging technology. In: Lassus P, Arzul G, Erard E, Gentien P, Marcaillou C (eds) Harmful marine algal blooms. Lavoiser Science Publishers, Paris, pp 3–13
Anderson DM, Walt DW (2009) A fiber optic microarray for the detection and enumeration of harmful algal bloom (HAB) species. Internal report: The NOAA/UNH cooperative institute for coastal and estuarine environmental technology (CICEET). http://ciceet.unh.edu/news/releases/spring09_reports/pdf/anderson_FR.pdf
Anderson DM, Kulis DM, Erdner D, Ahn S, Walt DR (2006) Fibre optic microarrays for the detection and enumeration of harmful algal bloom. Afr J Mar Sci 28(2):231–235
Andree KB, Fernández-Tejedor M, Elandaloussi LM, Quijano-Scheggia S, Sampedro N, Garcés E, Camp J, Diogéne J (2011) Quantitative PCR coupled with melt curve analysis for detection of selected Pseudo-nitzschia spp. (Bacillariophyceae) from the North Western Mediterranean sea. Appl Environ Microb 77(5):1651–1659
Anthony RM, Brown TJ, French GL (2000) Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J Clin Microbiol 38(2):781–788
Becker EW (2007) Microalgae as a source of protein. Biotenol Adv 25(2):207–210
Bertozzini E, PennaA PE, Bruce I, Magnani M (2005) Development of new procedures for the isolation of phytoplankton DNA from fixed samples. J Appl Phycol 17:223–229
Biegala IC, Kennaway G, Alverca E, Lennon JF, Vaulot D, Simon N (2002) Identification of bacteria associated with dinoflagellates (Dinophyceae) Alexandrium spp using tyramide signal amplification-fluorescent in situ hybridisation and confocal microscopy. J Phycol 38(2):404–411
Bott NJ, Ophel-Kellener KM, Sierp MT, Rowling KP, Mckay AC, Loo MGK, Tanner JE, Deveney MR (2010) Toward routine, DNA-based detection methods for marine pests. Biotechnol Adv 28:706–714
Bråte J, Klaveness D, Rygh T, Jakobsen KS, Shalchian-Tabrizi K (2010) Telonemia-specific environmental 18S rDNA PCR reveals unknown diversity and multiple marine-freshwater colonizations. BMC Microbiol 10:168. doi:10.1186/1471-2180-10-168
Britschgi TB, Giovannoni SJ (1991) Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl Environ Microbiol 57(6):1707–1713
Brown MR (2002) Nutritional value of microalgae for aquculture. In: Cruz-Suárez LE, Ricque-Marie D, Tapia-Salazar M, Gaxiola-Cortés MG, Simoes N (eds) Avances en Nutrición Acuícola VI. Memorias del VI Simposium Internacional de Nutrición Acuícola. 3 al 6 de Septiembre del 2002. Cancún, Quintana Roo, México
Burki F, Kudryavtsev A, Matz MV, Aglyamova GV, Bulman S, Fiers M, Keeling PJ, Pawlowski J (2010) Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists. BMC Evol Biol 10:377
Casper ET, Patterson SS, Bhanushali P, Farmer A, Smith M, Fries DP, Paul JH (2007) A handheld NASBA analyzer for the field detection and quantification of Karenia brevis. Harmful Algae 6(1):112–118
Casteleyn G, Leliaert F, Backeljau T, Debeer AE, Kotaki Y, Rhodes L, Lundholm N, Sabbe K, Vyverman W (2010) Limits to gene flow in a cosmopolitan marine planktonic diatom. Proc Natl Acad Sci USA 107(29):12952–12957
Cembella AD, Sullivan JJ, Boyer GL, Taylor FJR, Anderson RJ (1987) Variations in paralytic shellfish toxin composition within the Protogonyaulax tamarensis/catenella species complex: red tide dinoflagellates. Biochem Syst Ecol 15:171–186
Cheng KC, Ogden KL (2011) Algal biofuels: the research. American Institute of Chemical Engineers (AICHE). http://www.aiche.org
Cheung MK, Au CH, Chu KH, Kwan HS, Wong CK (2010) Composition and genetic diversity of pico eukaryotes in subtropical coastal waters as revealed by 454 pyrosequencing. ISME J 4:1053–1059
Chisti Y (2007) Biodiesel from microalgae. Biotechnol Adv 25(3):294–306
Cho SY, Nagai S, Nishitani G, Han MS (2009) Development of compound microsatellite markers in red-tide-causing dinoflagellate Akashiwo sanguinea (Dinophyceae). Mol Ecol Resour 9:915–917
Countway P, Gast RJ, Savala P, Caron DA (2005) Protistan diversity estimates based on 18S rDNA from seawater incubations in the Western North Atlantic. J Eukaryot Microbiol 52(2):95–106
de Bruin A, Ibelings BW, Donk EV (2003) Molecular techniques in phytoplankton research: from allozyme electrophoresis to genomics. Hydrobiologia 491(1–3):47–63
Delong EF, Pace NR (1991) Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol 173(14):4371–4378
Diaz MR, Jacobson JW, Goodwin KD, Dubar SA, Fell JW (2010) Molecular detection of harmful algal blooms (HABs) using locked nucleic acids and bead array technology. Limnol Oceanogr Methods 8:269–284
Díez B, Pedrós-Alió C, Marsh TL, Massana R (2001) Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine Pico eukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl Environ Microbiol 67(7):2942–2951
Dyhrman ST, Erdner D, La Du J, Galac M, Anderson DM (2006) Molecular quantification of toxic Alexandrium fundyense in the Gulf of Maine using real-time PCR. Harmful Algae 5(3):242–250
Dyhrman ST, Haley ST, Borchert JA, Lona B, Kollars N, Erdner DL (2010) Parallel analyses of Alexandrium catenella cell concentrations and shellfish toxicity in the Puget Sound. Appl Environ Microb 76(14):4647–4654
Edgcomb V, Orsi W, Bunge J, Jeon S, Christen R, Leslin C, Holder M, Taylor GT, Suarez P, Varela R, Epstein S (2011) Protistan microbial observatory in the Cariaco Basin, Caribbean I. pyrosequencing vs sanger insights in to species richness. ISME J 5:1344–1356. doi:10.1038/ismej.2011.6
Edvardsen B, Shalchian-Tabrizi K, Jakobsen KS, Medlin LK, Dahl E, Brubak S, Paasche E (2003) Genetic variability and molecular phylogeny of Dinophysis species (Dinophyceae) from Norwegian waters inferred from single cells analysis of rDNA. J Phycol 39(2):395–408
Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, Peterson DM, Saar MO, Alexander S, Alexander EC, Rohwer F (2006) Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 7:57
Ellison CK, Burton RS (2005) Application of bead array technology to community dynamics of marine phytoplankton. Mar Ecol Prog Ser 288:75–85
Elmqvist T, Folke C, Nyström M, Peterson G, Bengtsson J, Walker B, Norberg J (2003) Response diversity, ecosystem change, ecosystem change, and resilience. Fron Ecol Environ 1:488–494
Erdner DL, Percy L, Keafer B, Lewis J, Anderson DM (2010) A quantitative real-time PCR assay for the identification and enumeration of Alexandrium cysts in marine sediments. Deep Sea Res PT II 57:279–287
Evans KM, Chepurnov VA, Mann DG (2009) Ten microsatellite markers for the freshwater diatom Sellaphora capitata. Mol Ecol Resour 9:216–218
Galluzzi L, Penna A, Bertozzini E, Vila M, Garces E, Magnani M (2004) Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl Environ Microbiol 70(2):1199–1206
Galluzzi L, Bertozzini E, Penna A, Perini F, Garcés E, Magnani M (2010) Analysis of rRNA gene content in the Mediterranean dinoflagellate Alexandrium catenella and Alexandrium taylori: implications for the quantitative real-time PCR-based monitoring methods. J Appl Phycol 22(1):1–9
Galluzzi L, Cegna A, Casabianca S, Penna A, Saunders N, Magnani M (2011) Development of an oligonucleotide microarray for the detection and monitoring of marine dinoflagellates. J Microbiol Meth 84(2):234–242
Gescher C, Metfies K, Frickenhaus S, Knefelkamp B, Wiltshire KH, Medlin LK (2008) Feasibility of assessing the community composition of Prasinophytes at the Helgoland Roads sampling site with a DNA microarray. Appl Env Microbiol 74:5305–5316
Godhe A, Asplund ME, Härnström K, Saravanan V, Tyagi A, Karunasagar I (2008) Quantification of diatom and dinoflagellate biomasses in coastal marine seawater samples by real-time PCR. Appl Environ Microbiol 74(23):7174–7182
Guo Z, Guilfoyle RA, Thiel AJ, Wang R, Smith LM (1994) Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotides arrays on glass supports. Nucleic Acids Res 22:5456–5465
Handy SM, Demir E, Hutchins DA, Portune KJ, Whereat EB, Hare CE, Rose JM, Warner M, Farestad M, Cary S, Coyne KJ (2008) Using quantitative real-time PCR to study competition and community dynamics among Delaware Inland Bays harmful algae in field and laboratory studies. Harmful Algae 7(5):599–613
Hosoi-Tanabe S, Sako Y (2005) Rapid detection of natural cells of Alexandrium tamarense and A. catenella (Dinophyceae) by fluorescence in situ hybridization. Harmful Algae 4(2):319–328
Hosoi-Tanabe S, Sako Y (2006) Development and application of fluorescence in situ hybridization (FISH) method for simple and rapid identification of the toxic dinoflagellates Alexandrium tamarense and Alexandrium catenella in cultured and natural seawater. Fish Sci 72(1):77–82
Hubbard KA, Rocap G, Armbrust EV (2008) Inter- and intraspecific community structure within the diatom genus Pseudo-nitzschia (Bacillariophyceae). J Phycol 44(3):637–649
Joo S, Lee S-R, Park S (2010) Monitoring of phytoplankton community structure using terminal restriction fragment length polymorphism (T-RFLP). J Microbiol Meth 81(1):61–68
Ki J-S (2011) Hypervariable regions (V1-V9) of the dinoflagellate 18S rRNA using a large dataset for marker considerations. J Appl Phycol. doi:10.1007/s10811-011-9730-z
Ki J-S, Han M-S (2005) Sequence-based diagnostics and phylogenetic approach of uncultured freshwater dinoflagellate Peridinium (Dinophyceae) species, based on single-cell sequencing of rDNA. J Appl Phycol 17(2):147–153
Ki J-S, Han M-S (2006) A low-density oligonucleotide array study for parallel detection of harmful algal species using hybridization of consensus PCR products of LSU rDNA D2 domain. Biosens Bioelectron 21(9):1812–1821
Ki J-S, Jang GY, Han M-S (2004) Integrated method for single-cell DNA extraction, PCR amplification, and sequencing of ribosomal DNA from harmful dinoflagellates Cochlodinium polykrikoides and Alexandrium catenella. Mar Biotechnol 6(6):587–593
Kudela RM, Howard MDA, Jenkins BD, Miller PE, Smith GJ (2010) Using the molecular toolbox to compare harmful algal blooms in upwelling systems. Prog Oceanogr 85(1–2):108–121
Kumari N, Srivastava AK, Bhargava P, Rai LK (2009) Molecular approaches towards assessment of cyanobacterial biodiversity. Afr J Biotechnol 8(18):4284–4298
Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR (1985) Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA 82(20):6955–6959
Lepère C, Vaulot D, Scanlan DJ (2009) Photosynthetic picoeukaryotic community structure in the South East Pacific Ocean encompassing the most oligotrophic waters on Earth. Environ Microbiol 11(12):3105–3117
Lindquist HDA (1997) Probes for the specific detection of Cryptosporidium parvum. Water Res 31(10):2668–2671
Litaker RW, Tester PA (2006) Molecular approaches to the study of phytoplankton life cycles: implications for harmful algal bloom ecology. In: Granéli E, Turner T (eds) Ecology of harmful algae, ecological studies, vol 189. Springer, Heidelberg, pp 299–309
Liu W, Marsh T, Cheng H, Forney L (1997) Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol 63:4516–4522
Liu H, Wang H, Shi Z, Wang H, Yang C, Silke S, Tan W, Li Z (2006) TaqMan probe array for quantitative detection of DNA targets. Nucleic Acids Res 34(1):1–8
Man-Aharonovich D, Philosof A, Kirkup BC, Gal FL, Yogev T, Berman-Frank I, Polz MF, Vaulot D, Béjà O (2010) Diversity of active marine picoeukaryotes in the Eastern Mediterranean Sea unveiled using photosystem-II psbA transcripts. ISME J 4:1044–1052
Mao F, Leung W-Y, Xin X (2007) Characterization of EvaGreen and the implication of its physicochemical properties for qPCR applications. BMC Biotechnol 7:76
McCliment EA, Nelson CE, Carlson CA, Alldredge AL, Witting J, Amaral-Zettler LA (2011) An all-taxon microbial inventory of the Moorea coral reef ecosystem. ISME J. doi:10.1038/ismej.2011.108
Medinger R, Nolte V, Pandey RV (2010) Diversity in a hidden world: potential and limitation of next generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol Ecol 19(1):32–40
Medlin LK, Kooistra WHCF (2010) Methods to estimate the diversity in the marine photosynthetic protist community with illustrations from case studies: a review. Diversity 2:973–1014
Medlin LK, Lange M, Noethig EV (2000) Genetic diversity in the marine phytoplankton: a review and a consideration of Antarctic phytoplankton. Antarct Sc 12:325–331
Medlin LK, Metfies K, Mehl H, Wiltshire K, Valentin K (2006) Picoplankton diversity at the Helgoland Time Series Site as assessed by three molecular methods. Microb Ecol 167:1432–1451
Medlin LK, Metfies K, John U, Olsen J (2007) Algal molecular systematics: a review of the past and prospects for the future. In: Broadie J, Lewis J (eds) Unravelling the algae: the past, present and future of algal systematics. Sys Assn Special Vol Ser 75. CRC Press, Taylor & Francis Group, London, pp 341–353
Metfies K, Medlin LK (2008) Feasibility of transferring fluorescent in situ hybridization probes to an 18S rRNA gene phylochip and mapping of signal intensities. Appl Envron Microbiol 74:2814–2821
Metfies K, Huljic S, Lange M, Medlin LK (2005) Electrochemical detection of the toxic dinoflagellate Alexandrium ostenfeldii with a DNA-biosensor. Biosens Bioelectron 20:1349–1357
Metfies K, Töbe K, Scholin CA, Medlin LK (2006) Laboratory and field applications of ribosomal RNA probes to aid the detection and monitoring of Harmful Algae. In: Granéli E, Turner JT (eds) Ecology of harmful algae. Springer Verlag, Berlin, Heidelberg, pp 11–325
Metzker ML (2010) Sequencing technologies—the next generation. Nat Rev 11:31–46
Mitterer G, Huber M, Leidinger E, Kiristis C, Lubitz W, Mueller MW, Schmidt WM (2004) Microarray-based identification of bacteria in clinical samples by solid-phase PCR amplification of 23S ribosomal DNA sequences. J Clin Microbiol 42(2):1048–1057
Moreira D, López-García P (2002) The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol 10:31–38
Mori Y, Nagamine K, Tomita N, Notomi T (2001) Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Bioph Res Co 289:150–154
Nagai SC, Lian S, Yamaguchi M, Hamaguchi Y, Matsuyama S, Itakura H, Shimada S, Kaga H, Yamauchi Y, Sonda T, Kim C, Hogetsu T (2007) Microsatellite markers reveal population genetic structure of the toxic dinoflagellate Alexandrium tamarense (Dinophyceae) in Japanese coastal waters. J Phycol 43:43–54
Not F, Latasa M, Marie D, Cariou T, Vaulot D, Simon N (2004) A single species, Micromonas pusilla (Prasinophyceae), dominates the eukaryotic picoplankton in the Western English Channel. Appl Environ Microbiol 70(7):4064–4072
Not F, Gausling R, Azam F, Heidelberg JF, Worden AZ (2007) Vertical distribution of picoeukaryotic diversity in the open ocean. Environ Microbiol 9:1233–1252
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:E63
Nowrousian M (2010) Net-generation sequencing techniques for eukaryotic microorganisms: sequencing based solutions to biological problems. Eukaryot Cell 9(9):1300–1310
Oldach DW, Delwiche CF, Jakobsen KS, Tengs T, Brown EG, Kempton JW, Schaefer EF, Bowers HA, Glasgow HB Jr, Burkholder JM, Steidinger KA, Rublee PA (2000) Heteroduplex mobility assay-guided sequence discovery: elucidation of the small subunit (18S) rDNA sequences of Pfiesteria piscicida and related dinoflagellates from complex algal culture and environmental sample DNA pools. Proc Natl Acad Sci USA 97(8):4303–4308
Park TG, Salas MF, Bolch CJS, Hallegraeff GM (2007) Development of a realtime PCR probe for quantification of the heterotrophic dinoflagellate Cryptoperidiniopsis brodyi (Dinophyceae) in environmental samples. Appl Environ Microbiol 73:2552–2560
Park TG, Park YT, Lee Y (2009) Development of a SYTO9 based real-time PCR probe for detection and quantification of toxic dinoflagellate Karlodinium veneficum (Dinophyceae) in environmental samples. Phycologia 48(1):32–43
Penna A, Galluzzi L (2008) PCR techniques a diagnostic tool for the identification and enumeration of toxic marine phytoplankton species. In: Evangelista V, Barsanti L, Frassanito AM, Passarelli V, Gualtieri P (eds) Algal toxins: nature, occurrence, effect and detection. Springer Science + Business media BV, pp 261–284
Perini F, Casabianca A, Battocchi C, Accoroni S, Totti C, Penna A (2011) New approaches using the real-time PCR method for estimation of the toxic marine dinoflagellate Ostreopsis cf. ovata in marine environmental. PLoS One 6(3):e17699. doi:10.1371/journal.pone.0017699
Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic MD (2009) Metagenomic pyrosequencing and microbial identification. Clin Chem 55(5):856–866
Potvin M, Lovejoy C (2009) PCR-based diversity estimates of artificial and environmental 18s rRNA gene libraries. J Eukaryot Microbiol 56(2):174–181
Richlen ML, Barber PH (2005) A technique for the rapid extraction of microalgal DNA from single live and preserved cells. Mol Ecol Notes 5:688–691
Ripley SJ, Baker AS, Miller PI, Walne AW, Schroeder DC (2008) Development and validation of a molecular technique for the analysis of archived formalin-preserved phytoplankton samples permits retrospective assessment of Emiliania huxleyi communities. J Microbiol Meth. doi:10.1016/j.mimet.2008.02.001
Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, Daroub SH, Camargo FA, Farmerie WG, Triplett EW (2007) Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1(4):283–290
Roesch LFW, Lorca GL, Casella G, Giongo A, Naranjo A, Pionzio AM (2009) Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J 3:536–548
Rothberg JM, Leamon JH (2008) The development and impact of 454 sequencing. Nat Biotechnol 26:1117–1124
Rublee PA, Kempton JW, Schaefer EF, Allen C, Harris J, Oldach DW, Bowers H, Tengs T, Burkholder JM, Glasgow HB (2001) Use of molecular probes to assess geographic distribution of Pfiesteria species. Environ Health Perspect 109(5):765–767
Sako Y, Kim CH, Ninomiya H, Adachi M, Ishida Y (1990) Isozyme and cross analysis of mating populations in the Alexandrium catenella/tamarense species complex. In: Granéli E, Sundstrom B, Edler L, Anderson DM (eds) Toxic marine phytoplankton. Elsevier, New York, pp 320–323
Sarno D, Kooistra WHCF, Medlin LK, Percopo I, Zingone A (2005) Diversity in the genus Skeletonema (Bacillariophyceae): Skeletonema costatum (Bacillariophyceae) consists of several genetically and morphologically distinct species with the description of four new species. J Phycol 41:151–176
Schlötterer C (1998) Ribosomal DNA probes and primers. In: Karp A, Isaac PG, Ingram DS (eds) Molecular tools for screening biodiversity. Chapman & Hall, London, pp 267–276
Scholin CA, Anderson DA (1993) Population analysis of toxic and non-toxic Alexandrium species using ribosomal RNA signature sequences. In: Smayda TJ, Shimizu Y (eds) Toxic phytoplankton blooms in the sea. Elsevier, Amsterdam, pp 95–102
Scholin CA, Herzog M, Sogin ML, Anderson DM (1994) Identification of group and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). II. Sequence analysis of a fragment of the LSU rRNA gene. J Phycol 30:999–1011
Scorzetti G, Brand LE, Hitchcock GL, Rein KS, Sinigalliano CD, Fell JW (2009) Multiple simultaneous detection of harmful algal blooms (HABs) through a high throughput bead array technology, with potential use in phytoplankton community analysis. Harmful Algae 8:196–211
Sellner KG, Gregory E, Doucette J, Kirkpatric GJ (2003) Harmful algal blooms: causes, impacts and detection. J Ind Microbiol Biotechnol 30:383–440
Shalchian-Tabrizi K, Reier-Røberg K, Ree DK, Klaveness D, Brate J (2011) Marine-freshwater colonizations of haptophytes inferred from phylogeny of environmental 18S rDNA sequences. J Eukaryot Microbiol 58(11):315–318
Shapiro LP, Campbell L, Haugen EM (1989) Immunochemical recognition of phytoplankton species. Mar Ecol Prog Ser 57:219–224
Shendure J, Ji H (2008) Next generation DNA sequencing. Nat Biotechnol 26:1135–1145
Shi Y, Tyson GW, DeLong EF (2009) Metatranscriptomics reveals unique microbial small RNAs in the ocean’s water column. Nature 459(7244):266–269
Shi XL, Lepère C, Scanlan DJ, Vaulot D (2011) Plastid 16S rRNA gene diversity among eukaryotic picophytoplankton sorted by flow cytometry from the South Pacific Ocean. PLoS One 6(4):e18979. doi:10.1371/journal.pone.0018979
Simon N, Campbell L, Örnȯlfsdȯttir E, Groben R, Guillou L, Lange M, Medlin LK (2000) Oligonucleotide probes for the identification of three algal groups by dot blot and Fluorescent Whole-Cell Hybridization. J Eukaryot Microbiol 47(1):76–84
Soto K, Collantes G, Zahr M, Kuzhar J (2005) Simultaneous enumeration of Phaeodactylum tricornutum (MCB292) and bacteria growing in mixed communities. Invest Mar Valparaiso 33(2):143–149
Stackebrandt E (2006) Molecular identification, systematics, and population structure of prokaryotes. Springer-Verlag Berlin Heidelberg, Germany, pp 51–80
Stoeck T, Epstein S (2003) Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl Environ Microbiol 69(5):2657–2663
Stoeck T, Bass D, Nebel M, Christen R, Jones MD, Breiner HW, Richard TA (2010) Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol 19(1):21–31
Tai V, Poon AF, Paulsen IT, Palenik B (2011) Selection in coastal Synechococcus (cyanobacteria) populations eva;uated from environmental metagenomics. PLoS One 6(9):e24249
Takano Y, Horiguchi T (2006) Acquiring scanning electron microscopical. Light microscopical and multiple gene sequence data from a single dinoflagellate cell. J Phycol 42:251–256
Töbe K, Eller G, Medlin LK (2006) Automated detection and enumeration for toxic algae by solid-phase cytometry and the introduction of a new probe for Prymnesium parvum (Haptophyta:Prymnesiophyceae). J Plank Res 28(7):643–657
Toyoda K, Nagasaki K, Tomaru Y (2010) Application of real-time PCR assay for detection and quantification of bloom-forming diatom Chaetoceros tenuissimus Meunier. Plankton Benthos Res 5(2):56–61
Tyrrell JV, Connell LB, Scholin CA (2002) Monitoring for Heterosigma akashiwo using a sandwich hybridization assay. Harmful Algae 1:205–214
Ulrich RM, Casper ET, Campbell L, Richardson B, Heil CA, Paul JH (2010) Detection and quantification of Karenia mikimotoi using real-time nucleic acid sequence-based amplification with internal control RNA (IC-NASBA). Harmful Algae 9(1):116–122
Vaulot D, Eikrem W, Viprey M, Moreau H (2008) The diversity of small eukaryotic phytoplankton (3 mm) in marine ecosystems. FEMS Microbiol Rev 32:795–820
Wang L, Li MJ, Alam Y, Geng Z, Yamasaki S, Shi L (2008) Loop-mediated isothermal amplification method for rapid detection of the toxic dinoflagellate Alexandrium, which causes algal blooms and poisoning of shellfish. FEMS Microbiol Lett 282:15–21
Widmer F, Hartmann M, Frey B, Kölliker B (2006) A novel strategy to extract specific phylogenetics sequence information from community T-RFLP. J Microbiol Meth 66:512–520
Wu Z, Irizarry RA, Gentleman R, Murillo FM, Spencer F (2003) A model based background adjustment for oligonucleotide expression arrays. Technical report, Johns Hopkins University, dept of biostatistics working papers. http://www.bepress.com/jhubiostat/paper1
Yershov G, Barsky V, Kirillov E, Kreindlin K, Ivanov I, Parinov S, Guschin D, Drobishev A, Dubiley S (1996) DNA analysis and diagnostics on oligonucleotide microchips. Proc Natl Acad Sci USA 93(10):4913–4918
Yoon HS, Price DC, Stepanauskas R, Rajah VD, Sieracki ME, Wilson WH, Yang EC, Duffy S, Bhattacharya D (2011) Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 332:714–717
Zhu F, Massana R, Not F, Marie D, Vaulot D (2005) Mapping of picoeukaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol 52:79–92
Acknowledgments
This work was supported by both the Marine and Extreme Genome Research Center Program of the Ministry of Land, Transportation and Maritime Affairs, Republic of Korea, and by the National Research Foundation of Korea Grant funded by the Korean Government (MEST) (NRF-C1ABA001-2011-0018573).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ebenezer, V., Medlin, L.K. & Ki, JS. Molecular Detection, Quantification, and Diversity Evaluation of Microalgae. Mar Biotechnol 14, 129–142 (2012). https://doi.org/10.1007/s10126-011-9427-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-011-9427-y