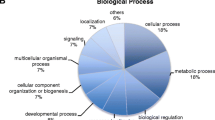
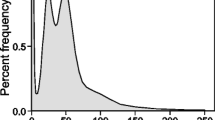
Abstract
DNA fingerprints and end sequences from bacterial artificial chromosomes (BACs) from two new libraries were generated to improve the first generation integrated physical and genetic map of the rainbow trout (Oncorhynchus mykiss) genome. The current version of the physical map is composed of 167,989 clones of which 158,670 are assembled into contigs and 9,319 are singletons. The number of contigs was reduced from 4,173 to 3,220. End sequencing of clones from the new libraries generated a total of 11,958 high quality sequence reads. The end sequences were used to develop 238 new microsatellites of which 42 were added to the genetic map. Conserved synteny between the rainbow trout genome and model fish genomes was analyzed using 188,443 BAC end sequence (BES) reads. The fractions of BES reads with significant BLASTN hits against the zebrafish, medaka, and stickleback genomes were 8.8%, 9.7%, and 10.5%, respectively, while the fractions of significant BLASTX hits against the zebrafish, medaka, and stickleback protein databases were 6.2%, 5.8%, and 5.5%, respectively. The overall number of unique regions of conserved synteny identified through grouping of the rainbow trout BES into fingerprinting contigs was 2,259, 2,229, and 2,203 for stickleback, medaka, and zebrafish, respectively. These numbers are approximately three to five times greater than those we have previously identified using BAC paired ends. Clustering of the conserved synteny analysis results by linkage groups as derived from the integrated physical and genetic map revealed that despite the low sequence homology, large blocks of macrosynteny are conserved between chromosome arms of rainbow trout and the model fish species.




Similar content being viewed by others
References
Allendorf FW, Thorgaard GH (1984) Tetraploidy and the evolution of salmonid fishes. In: Turner BJ (ed) Evolutionary genetics of fishes. Plenum, New York
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Baerwald MR, Petersen JL, Hedrick RP, Schisler GJ, May B (2010) A major effect quantitative trait locus for whirling disease resistance identified in rainbow trout (Oncorhynchus mykiss). Heredity 106:920–926
Barroso RM, Wheeler PA, Lapatra SE, Drew RE, Thorgaard GH (2008) QTL for IHNV resistance and growth identified in a rainbow (Oncorhynchus mykiss)xYellowstone cutthroat (Oncorhynchus). Aquaculture 277:156–163
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF (2001) M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. Biotechniques 31(24–6):28
Cole J, Wiggans G, Ma L, Sonstegard T, Lawlor T, Crooker B, Van Tassell C, Yang J, Wang S, Matukumalli L, Da Y (2011) Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows. BMC Genomics 12:408
Dalrymple B, Kirkness E, Nefedov M, Mcwilliam S, Ratnakumar A, Barris W, Zhao S, Shetty J, Maddox J, O’grady M, Nicholas F, Crawford A, Smith T, De Jong P, Mcewan J, Oddy VH, Cockett N, The International Sheep Genomics C (2007) Using comparative genomics to reorder the human genome sequence into a virtual sheep genome. Genome Biol 8:R152
Danzmann RG, Cairney M, Davidson WS, Ferguson MM, Gharbi K, Guyomard R, Holm LE, Leder E, Okamoto N, Ozaki A, Rexroad CE, Sakamoto T, Taggart JB, Woram RA (2006) A comparative analysis of the rainbow trout genome with 2 other species of fish (Arctic charr and Atlantic salmon) within the tetraploid derivative Salmonidae family (subfamily: Salmoninae). Genome 48:1037–1051
Danzmann R, Davidson E, Ferguson M, Gharbi K, Koop B, Hoyheim B, Lien S, Lubieniecki K, Moghadam H, Park J, Phillips R, Davidson W (2008) Distribution of ancestral proto-Actinopterygian chromosome arms within the genomes of 4R-derivative salmonid fishes (Rainbow trout and Atlantic salmon). BMC Genomics 9:557
Davidson W, Koop B, Jones S, Iturra P, Vidal R, Maass A, Jonassen I, Lien S, Omholt S (2011) Sequencing the genome of the Atlantic salmon (Salmo salar). Genome Biol 11:403
Drew RE, Schwabl H, Wheeler PA, Thorgaard GH (2007) Detection of QTL influencing cortisol levels in rainbow trout (Oncorhynchus mykiss). Aquaculture 272:S183–S194
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Falgueras J, Lara A, Fernandez-Pozo N, Canton F, Perez-Trabado G, Claros MG (2010) SeqTrim: a high-throughput pipeline for pre-processing any type of sequence read. BMC Bioinforma 11:38
Genet C, Dehais P, Palti Y, Gao G, Gavory F, Wincker P, Quillet E, Boussaha M (2011) Analysis of BAC-end sequences in rainbow trout: content characterization and assessment of synteny between trout and other fish genomes. BMC Genomics 12:314
Govoroun M, Le Gac F, Guiguen Y (2006) Generation of a large scale repertoire of expressed sequence tags (ESTs) from normalised rainbow trout cDNA libraries. BMC Genomics 7:196
Guyomard R, Mauger S, Tabet-Canale K, Martineau S, Genet C, Krieg F, Quillet E (2006) A type I and type II microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) with presumptive coverage of all chromosome arms. BMC Genomics 7:302
Haidle L, Janssen J, Gharbi K, Moghadam H, Ferguson M, Danzmann R (2008) Determination of quantitative trait loci (QTL) for early maturation in rainbow trout (Oncorhynchus mykiss). Mar Biotechnol 10:579–592
Katagiri T, Asakawa S, Minagawa S, Shimizu N, Hirono I, Aoki T (2001) Construction and characterization of BAC libraries for three fish species; rainbow trout, carp and tilapia. Anim Genet 32:200–204
Kuhl H, Beck A, Wozniak G, Canario A, Volckaert F, Reinhardt R (2010) The European sea bass Dicentrarchus labrax genome puzzle: comparative BAC-mapping and low coverage shotgun sequencing. BMC Genomics 11:68
Kuhl H, Sarropoulou E, Tine M, Kotoulas G, Magoulas A, Reinhardt R (2011) A comparative BAC map for the gilthead sea bream (Sparus aurata L.). J Biomed Biotechnol 2011:1–7
Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci U S A 84:2363–2367
Larkin DM, Wind A, Rebeiz M, Schweitzer PA, Bachman S, Green C, Wright CL, Campos EJ, Benson LD, Edwards J, Liu L, Osoegawa K, Womack JE, De Jong PJ, Lewin HA (2003) A cattle–human comparative map built with cattle BAC-ends and human genome sequence. Genome Res 13:1966–1972
Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci U S A 81:3443–3446
Le Bras Y, Dechamp N, Krieg F, Filangi O, Guyomard R, Boussaha M, Bovenhuis H, Pottinger T, Prunet P, Le Roy P, Quillet E (2011) Detection of QTL with effects on osmoregulation capacities in the rainbow trout (Oncorhynchus mykiss). BMC Genet 12:46
Liu H, Jiang Y, Wang S, Ninwichian P, Somridhivej B, Xu P, Abernathy J, Kucuktas H, Liu Z (2009) Comparative analysis of catfish BAC end sequences with the zebrafish genome. BMC Genomics 10:592
Luo M-C, Thomas C, You FM, Hsiao J, Ouyang S, Buell CR, Malandro M, Mcguire PE, Anderson OD, Dvorak J (2003) High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics 82:378–389
Matise TC, Perlin M, Chakravarti A (1994) Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat Genet 6:384–390
Miller MR, Brunelli JP, Wheeler PA, Liu S, Rexroad Iii CE, Palti Y, Doe CQ & Thorgaard GH (2011) A conserved haplotype controls parallel adaptation in geographically distant salmonid populations. Molecular Ecology. doi:10.1111/j.1365-294X.2011.05305.x
Nelson W, Soderlund C (2009) Integrating sequence with FPC fingerprint maps. Nucl Acids Res 37:e36
Nelson WM, Bharti AK, Butler E, Wei F, Fuks G, Kim H, Wing RA, Messing J, Soderlund C (2005) Whole-genome validation of high-information-content fingerprinting. Plant Physiol 139:27–38
Ng SH, Artieri CG, Bosdet IE, Chiu R, Danzmann RG, Davidson WS, Ferguson MM, Fjell CD, Hoyheim B, Jones SJ, De Jong PJ, Koop BF, Krzywinski MI, Lubieniecki K, Marra MA, Mitchell LA, Mathewson C, Osoegawa K, Parisotto SE, Phillips RB, Rise ML, Von Schalburg KR, Schein JE, Shin H, Siddiqui A, Thorsen J, Wye N, Yang G, Zhu B (2005) A physical map of the genome of Atlantic salmon, Salmo salar. Genomics 86:396–404
Nichols KM, Bartholomew J, Thorgaard GH (2003a) Mapping multiple genetic loci associated with Ceratomyxa shasta resistance in Oncorhynchus mykiss. Dis Aquat Organ 56:145–154
Nichols KM, Young WP, Danzmann RG, Robison BD, Rexroad C, Noakes M, Phillips RB, Bentzen P, Spies I, Knudsen K (2003b) A consolidated linkage map for rainbow trout (Oncorhynchus mykiss). Anim Genet 34:102–115
Nichols KM, Broman KW, Sundin K, Young JM, Wheeler PA, Thorgaard GH (2007) Quantitative trait loci × maternal cytoplasmic environment interaction for development rate in Oncorhynchus mykiss. Genetics 175:335–347
Nichols KM, Edo AF, Wheeler PA, Thorgaard GH (2008) The genetic basis of smoltification-related traits in Oncorhynchus mykiss. Genetics 179:1559–1575
Nievergelt CM, Smith DW, Kohlenberg JB, Schork NJ (2004) Large-scale integration of human genetic and physical maps. Genome Res 14:1199–1205
O’connell JR, Weeks DE (1995) The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet 11:402–408
O’connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266
O’malley KG, Sakamoto T, Danzmann RG, Ferguson MM (2003) Quantitative trait loci for spawning date and body weight in rainbow trout: testing for conserved effects across ancestrally duplicated chromosomes. J Hered 94:273–284
Ozaki A, Sakamoto T, Khoo S, Nakamura K, Coimbra MR, Akutsu T, Okamoto N (2001) Quantitative trait loci (QTLs) associated with resistance/susceptibility to infectious pancreatic necrosis virus (IPNV) in rainbow trout (Oncorhynchus mykiss). Mol Genet Genomics 265:23–31
Palti Y, Gahr SA, Hansen JD, Rexroad CE (2004) Characterization of a new BAC library for rainbow trout: evidence for multi-locus duplication. Anim Genet 35:130–133
Palti Y, Luo M-C, Hu Y, Genet C, You F, Vallejo R, Thorgaard G, Wheeler P, Rexroad C (2009) A first generation BAC-based physical map of the rainbow trout genome. BMC Genomics 10:462
Palti Y, Genet C, Luo M-C, Charlet A, Gao G, Hu Y, Castano-Sanchez C, Tabet-Canale K, Krieg F, Yao J, Vallejo R, Rexroad Iii C (2011) A first generation integrated map of the rainbow trout genome. BMC Genomics 12:180
Perry GM, Danzmann RG, Ferguson MM, Gibson JP (2001) Quantitative trait loci for upper thermal tolerance in outbred strains of rainbow trout (Oncorhynchus mykiss). Heredity 86:333–341
Perry GM, Ferguson MM, Sakamoto T, Danzmann RG (2005) Sex-linked quantitative trait loci for thermotolerance and length in the rainbow trout. J Hered 96:97–107
Phillips RB, Nichols KM, Dekoning JJ, Morasch MR, Keatley KA, Rexroad C 3rd, Gahr SA, Danzmann RG, Drew RE, Thorgaard GH (2006) Assignment of rainbow trout linkage groups to specific chromosomes. Genetics 174:1661–1670
Phillips R, Keatley K, Morasch M, Ventura A, Lubieniecki K, Koop B, Danzmann R, Davidson W (2009) Assignment of Atlantic salmon (Salmo salar) linkage groups to specific chromosomes: conservation of large syntenic blocks corresponding to whole chromosome arms in rainbow trout (Oncorhynchus mykiss). BMC Genet 10:46
Quillet E, Dorson M, Le Guillou S, Benmansour A, Boudinot P (2007) Wide range of susceptibility to rhabdoviruses in homozygous clones of rainbow trout. Fish Shellfish Immunol 22:510–519
Quiniou S, Waldbieser G, Duke M (2007) A first generation BAC-based physical map of the channel catfish genome. BMC Genomics 8:40
Rexroad CE, Lee Y, Keele JW, Karamycheva S, Brown G, Koop B, Gahr SA, Palti Y, Quackenbush J (2003) Sequence analysis of a rainbow trout cDNA library and creation of a gene index. Cytogenet Genome Res 102:347–354
Rexroad CE 3rd, Palti Y, Gahr SA, Vallejo RL (2008) A second generation genetic map for rainbow trout (Oncorhynchus mykiss). BMC Genet 9:74
Rise ML, Von Schalburg KR, Brown GD, Mawer MA, Devlin RH, Kuipers N, Busby M, Beetz-Sargent M, Alberto R, Gibbs AR, Hunt P, Shukin R, Zeznik JA, Nelson C, Jones SRM, Smailus DE, Jones SJM, Schein JE, Marra MA, Butterfield YSN, Stott JM, Ng SHS, Davidson WS, Koop BF (2004) Development and application of a salmonid EST database and cDNA microarray: data mining and interspecific hybridization characteristics. Genome Res 14:478–490
Robison BD, Wheeler PA, Thorgaard GH (1999) Variation in development rate among clonal lines of rainbow trout (Oncorhynchus mykiss). Aquaculture 173:131–141
Robison BD, Wheeler PA, Sundin K, Sikka P, Thorgaard GH (2001) Composite interval mapping reveals a major locus influencing embryonic development rate in rainbow trout (Oncorhynchus mykiss). J Hered 92:16–22
Rodriguez MF, Lapatra S, Williams S, Famula T, May B (2004) Genetic markers associated with resistance to infectious hematopoietic necrosis in rainbow and steelhead trout. Aquaculture 241:93–115
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Sakamoto T, Danzmann RG, Okamoto N, Ferguson MM, Ihssen PE (1999) Linkage analysis of quantitative trait loci associated with spawning time in rainbow trout (Oncorhynchus mykiss). Aquaculture 173:33–43
Sakamoto T, Danzmann RG, Gharbi K, Howard P, Ozaki A, Khoo SK, Woram RA, Okamoto N, Ferguson MM, Holm L-E, Guyomard R, Hoyheim B (2000) A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155:1331–1345
Salem M, Kenney B, Rexroad CE, Yao J (2008) Development of a 37 k high-density oligonucleotide microarray: a new tool for functional genome research in rainbow trout. J Fish Biol 72:2187–2206
Salem M, Xiao C, Womack J, Rexroad CE, Yao J (2009) MicroRNA repertoire for functional genome research in rainbow trout (Oncorhynchus mykiss). Marine Biotech 12:410–429
Salem M, Rexroad C, Wang J, Thorgaard G, Yao J (2010) Characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches. BMC Genomics 11:564
Soderlund C, Longden I, Mott R (1997) FPC: a system for building contigs from restriction fingerprinted clones. Comput Appl Biosci 13:523–535
Soderlund C, Humphray S, Dunham A, French L (2000) Contigs built with fingerprints, markers, and FPC V4.7. Genome Res 10:1772–1787
Sundin K, Brown KH, Drew RE, Nichols KM, Wheeler PA, Thorgaard GH (2005) Genetic analysis of a development rate QTL in backcrosses of clonal rainbow trout, Oncorhynchus mykiss. Aquaculture 247:75–83
Thorgaard GH, Bailey GS, Williams D, Buhler DR, Kaattari SL, Ristow SS, Hansen JD, Winton JR, Bartholomew JL, Nagler JJ, Walsh PJ, Vijayan MM, Devlin RH, Hardy RW, Overturf KE, Young WP, Robison BD, Rexroad CE, Palti Y (2002) Status and opportunities for genomics research with rainbow trout. Comp Biochem Physiol B Biochem Mol Biol 133:609–646
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wiggans GR, Sonstegard TS, Vanraden PM, Matukumalli LK, Schnabel RD, Taylor JF, Schenkel FS, Van Tassell CP (2009) Selection of single-nucleotide polymorphisms and quality of genotypes used in genomic evaluation of dairy cattle in the United States and Canada. J Dairy Sci 92:3431–3436
Xu P, Wang S, Liu L, Peatman E, Somridhivej B, Thimmapuram J, Gong G, Liu Z (2006) Channel catfish BAC-end sequences for marker development and assessment of syntenic conservation with other fish species. Anim Genet 37:321–326
Xu P, Li J, Li Y, Cui R, Wang J, Wang J, Zhang Y, Zhao Z, Sun X (2011) Genomic insight into the common carp (Cyprinus carpio) genome by sequencing analysis of BAC-end sequences. BMC Genomics 12:188
Young WP, Wheeler PA, Fields RD, Thorgaard GH (1996) DNA fingerprinting confirms isogenicity of androgenetically derived rainbow trout lines. J Hered 87:77–80
Young WP, Wheeler PA, Coryell VH, Keim P, Thorgaard GH (1998) A detailed linkage map of rainbow trout produced using doubled haploids. Genetics 148:839–850
Zimmerman A, Evenhuis J, Thorgaard G, Ristow S (2004) A single major chromosomal region controls natural killer cell-like activity in rainbow trout. Immunogenetics 55:825–835
Acknowledgments
This project was supported by National Research Initiative Competitive Grant number 2007-35616-17875 from the USDA National Institute of Food and Agriculture and by internal base funds provided by the Agricultural Research Service project number 1930-31000-009. Brian Smith, Kristy Shewbridge, and Roseanna Long carried out the microsatellites genotyping described in this manuscript. We thank Gary Thorgaard and Paul Wheeler (Washington State University) for the Swanson doubled haploid samples that were used to construct the new BAC libraries and Eric Peatman (Auburn University) for reviewing an earlier draft of the manuscript. We also thank anonymous reviewer 2 for his insightful comments on the blocks of conserved macro-synteny that can be identified through the chromosomes of the integrated map. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
File 1
Microsatellite markers information in Excel file. (XLS 32 kb)
File 2
Genetic map data and statistics in excel sheets. (XLS 208 kb)
File 3
A schematic drawing of the genetic map linkage groups. (PDF 5331 kb)
File 4
Detailed results of the BlastN conserved synteny analyses in excel sheets. (XLS 868 kb)
File 5
Detailed results of the BlastX conserved synteny analyses in excel sheets. (XLS 741 kb)
File 6
Statistics of BlastN and BlastX hits on all three species. (XLS 19 kb)
Rights and permissions
About this article
Cite this article
Palti, Y., Genet, C., Gao, G. et al. A Second Generation Integrated Map of the Rainbow Trout (Oncorhynchus mykiss) Genome: Analysis of Conserved Synteny with Model Fish Genomes. Mar Biotechnol 14, 343–357 (2012). https://doi.org/10.1007/s10126-011-9418-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-011-9418-z