Abstract
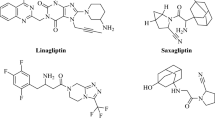
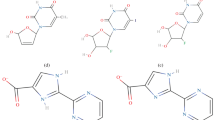
We apply molecular docking, molecular dynamics (MD) simulation, and binding free energy calculation to investigate and reveal the binding mechanism between five xanthine inhibitors and DPP-4. The electrostatic and van der Waals interactions of the five inhibitors with DPP-4 are analyzed and discussed. The computed binding free energies using MM-PBSA method are in qualitatively agreement with experimental inhibitory potency of five inhibitors. The hydrogen bonds of inhibitors with Ser630 and Asp663 can stabilize the inhibitors in binding sites. The van der Waals interactions, especially the key contacts with His740, Asn710, Trp629, and Tyr666 have larger contributions to the binding free energy and play important roles in distinguishing the variant bioactivity of five inhibitors.








Similar content being viewed by others
References
Feng J, Zhang Z, Michael B (2007) Discovery of alogliptin: a potent, selective, bioavailable, and efficacious inhibitor of dipeptidyl peptidase IV. J Med Chem 50:2297–2300
Wright S, Mark J (2006) cis-2,5-Dicyanopyrrolidine inhibitors of dipeptidyl peptidase IV: synthesis and in vitro, in vivo, and x-ray crystallographic characterization. J Med Chem 49:3068–3076
Rosenstock J, Schwartz SL, Clark CM Jr, Park GD, Donley DW, Edwards MB (2001) Basal insulin therapy in type 2 diabetes: 28-week comparison of insulin glargine (HOE 901) and NPH insulin. Diabetes Care 24:631–636
Barnett AH, Owens DR (1997) Insulin analogues. Lancet 349:47–51
King P, Peacock I, Donnelly R (1999) The UK Prospective Diabetes Study (UKPDS): clinical and therapeutic implications for type 2 diabetes. Br J Clin Pharmacol 48:643–648
UK Prospective Diabetes Study Group (1990) UK prospective diabetes study 7: response of fasting plasma glucose to diet therapy in newly presenting type II diabetic patients. Metabolism 39:905–912
Gerich JE (1989) Oral hypoglycemic agents. N Engl J Med 321:1231–1245
Lillelund H, Jensen H, Liang X, Bols M (2002) Recent developments of transition-state analogue glycosidase inhibitors of non-natural product origin. Chem Rev 102:515–553
Berecibar A, Grandjean C, Siriwardena A (1999) Synthesis and biological activity of natural aminocyclopentitol glycosidase inhibitors: mannostatins, trehazolin, allosamidins, and their analogs. Chem Rev 99:779–844
Jacob S (1995) Glycosylation inhibitors in biology and medicine. Curr Opin Struct Biol 5:605–611
Saltiel R, Olefsky M (1996) Thiazolidinediones in the treatment of insulin resistance and type II diabetes. Diabetes 45:1661–1669
Schoonjans K, Martin G, Staels B, Auwerx J (1997) Peroxisome proliferator-activated receptors, or-phans with ligands and functions. Curr Opin Lipidol 8:159–166
Pittas G, Greenberg S (2002) Thiazolidinediones in the treatment of type 2 diabetes. Expert Opin Pharmacother 3:529–540
Rami K, Smith A (2000) Synthetic ligands for PPARç s review of patent literature 1994–1999. Expert Opin Ther Pat 10:623–634
Van Gaal L, Scheen J (2002) Are all glitazones the same diabetes. Metab Res Rev 18:S1–S4
Hauner H (2002) The mode of action of thiazolidinediones. Diabetes Metab Res Rev 18:S10–S15
Witters L (2001) The blooming of the French lilac. J Clin Invest 108:1105–1107
Bailey J, Path C, Turner C (1996) Drug therapy-metformin N. Engl J Med 334:574–579
Wagman AS, Nuss JM (2001) Current therapies and emerging targets for the treatment of diabetes. Curr Pharm Des 7:417–450
Deacon CF (2011) Dipeptidyl peptidase-4 inhibitors in the treatment of type 2 diabetes:a comparative review. Diabetes Obes Metab 13:7–18
Drucker DJ (2001) Minireview: the glucagon-like peptides. Endocrinology 142:521–527
Ross SA, Gulve EA, Wang M (2004) Chemistry and biochemistry of type 2 diabetes. Chem Rev 104:1255–1282
Bo A (2009) Islet G protein-coupled receptors as potential targets for treatment of type 2 diabetes. Nat Rev Drug Discov 8:369–385
Mojsov S, Weir GC, Habener JF (1987) Insulinotropin: Glucagon-like peptide I (7–37) co-encoded in the glucagon gene is a potentstimulator of insulin release in the perfused rat pancreas. J Clin Invest 79:616–619
Kreymann B, Ghatei MA, Williams G, Bloom SR (1987) Glucagon-like peptide-1 7–36: a physiological incretin in man. Lancet 2:1300–1304
Rskov C, Holst JJ, Nielsen OV (1988) Effect of truncated glucagon-like peptide-1 [proglucagon-(78–107) amide] on endocrine secretionfrom pig pancreas, antrum, and nonantral stomach. Endocrinology 123:2009–2013
Nauck MA, Heimesaat MM, Behle K (2002) Effects of glucagon- like peptide 1 on counterregulatory hormone responses, cognitive functions, and insulin secretion during hyperinsulinemic, stepped hypoglycemic clamp experiments in healthy volunteers. J Clin Endocrinol Metab 87:1239–1246
Wettergren A, Schjoldager B, Mortensen PE (1993) Truncated GLP-1 (proglucagon 78–107-amide) inhibits gastric and pancreatic functions in man. Dig Dis Sci 38:665–673
Nauck MA, Niedereichholz U, Ettler R, Holst JJ, Orskov C, Ritzel R, Schmiegel WH (1997) Glucagon-like peptide 1 inhibition of gastric emptying outweighs its insulino-tropic effects in healthy humans. Am J Physiol 273:E981–E988
Flint A, Raben A, Ersboll AK, Holst JJ, Astrup A (2001) The effect of physiological levels of glucagons-like peptide-1 on appetite, gastric emptying, energy and substrate metabolism in obesity. Int J Obes 25:781–792
Magnin DR, Robl JA (2004) Synthesis of novel potent dipeptidyl peptidase IV inhibitors with enhanced chemical stability: interplay between the N-terminal amino acid alkyl side chain and the cyclopropyl group of α-aminoacyl-L-cis-4,5-methanoprolinenitrile-based inhibitors. J Med Chem 47:2587–2598
Hiramatsu H, Yamamoto A, Kyono K (2004) The crystal structure of human dipeptidyl peptidase IV(DPPIV) complex with diprotin. A Biol Chem 385(6):561–564
Lambei AM, Seharpe, DeMeester (2008) DPP-4 inhibitors for diabetes- what next. Biochem Pharmacol 76(12):1637–1643
Mentlein R, Gallwitzand B, Schmidt WE (1993) Dipeptidyl-peptidlase IV hydrolyses gastric inhibitory polypeptide, glueagon- like PePtide- l (7–36) amide peptide histidine methionine and 15 responsible for their degradation in human serum. EurJBioehem 214(3):829–835
Chien C-H, Tsai C-H, Lin C-H (2006) Identification of hydrophobic residues critical for DPP-IV dimerization. Biochemistry 45(23):7006–7012
Hansen L, Deacon CF, Orskov C, Holst JJ (1999) Glucagon- like peptide-1 (7–36) amide is transformed to glucagon-like peptide-1 (9–36) amide by dipeptidyl peptidase IV in the capillaries supplying the L-cells of the porcine intestine. Endocrinology 140:5356–5363
Kieffer TJ, McIntosh CHS, Pederson RA (1995) Degradation of glucose-dependent insulinotropic polypeptide and truncated glucagon-like Pep- tide-1 in vitro and in vivo by dipeptidyl peptidase IV. Endocrinology 136:3585–3597
Gwaltney SL, Stafford JA (2005) Inhibitors of dipeptidyl peptidase. Annu Rep Med Chem 40:149–165
Hiramatsu H, Yamamoto A, Kyono K (2004) The crystal structure of human dipeptidyl peptidase IV(DPPIV) complex with diprotin A. Biol Chem 385:561–564
Chien C-H, Tsai C-H, Lin C-H (2006) Identification of hydrophobic residues critical for DPP-IV dimerization. Biochemistry 45:7006–7012
Engel M, Hoffmann T, Manhart S (2006) Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV. J Mol Biol 355:768–783
Barnett A (2006) DPP-4 inhibitors and their potential role in the management of type 2 diabetes. Int J Clin Pract 60:1454–1470
Eckhardt M, Langkopf E, Mark M (2007) 8-(3-(R)-aminopiperidin-1-yl)-7-but-2-ynyl-3-methyl-1-(4-methyl-quinazolin-2-ylmethyl)-3,7-dihydropurine-2,6-dione (BI 1356), a highly potent, selective, long-acting, and orally bioavailable DPP-4 inhibitor for the treatment of type 2 diabetes. J Med Chem 50:6450–6453
Li C, Weiqiang L, Chunhua L (2012) Identification of diverse dipeptidyl peptidase IV inhibitors via structure-based virtual screening. J Mol Model 18:4033–4042
Li C, Shen J, Li W (2011) Possible ligand release pathway of dipeptidyl peptidase IV investigated by molecular dynamics simulations. Wiley-Liss Inc 79:1800–1809
Pradip Jadav, Rajesh Bahekar, Shailesh R. Shah (2012) Long-acting peptidomimetics based DPP-IV inhibitors. Bioorg Med Chem Lett
Janey J, Hsiao Y, Joseph Armstrong III (2006) Proline-catalyzed, asymmetric mannich reactions in the synthesis of a DPP-IV inhibitor. J Org Chem 71:390–392
Green BD, Flatt PR, Bailey CJ (2006) Dipeptidyl peptidase IV (DPP IV) inhibitors: a newly emerging drug class for the treatment of type 2 diabetes. Vasc Dis Res 3:159–165
Gue Hao, Yanzheng Cai, Weicheng Zhou (2009) Progress in the research of dipeptidyl peptidase-4 inhibitors. World Clinical Drugs 30 NO.8
Liang D, Gao J, Cheng Y, Cui W, Zhang H, Ji M (2011) Molecular dynamics simulations and MM/GBSA methods to investigate binding mechanisms of amino methyl pyrimidine inhibitors with DPP-IV. Bioorg Med Chem Lett 21:6630–6635
Wei C, Desheng L, Jian G, Fang L, Lingling G, Mingjuan J (2013) Molecular dynamics and free energy studies of chirality specificity effects on aminobenzo[a]quinolizine inhibitors binding to DPP-IV. J Mol Model 19:1167–1177
Otyepka M, Skopalík J (2007) What common structural features and variations of mammalian P450s are known to date? Biochim Biophys Acta 1770:376–389
Vincent BC, Tan BZ, Lim KM, Tay TE (2010) Explaining the inhibition of cyclin-dependent kinase 5 by peptides derived from p25 with molecular dynamics simulations and MM-PBSA. J Mol Model 16:1–8
Chen Q, Cui W, Ji M (2009) Studies of chirality effect of 4-(phenylamino)-pyrrolo[2,1-f] [1,2,4]triazine on p38a by molecular dynamics simulations and free energy calculations. J Comput Aided Mol Des 23:737–745
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB Ortiz JV, Cioslowski J, Fox DJ (2009) Gaussian 09 Revision.A.02. Gaussian Inc, Wallingford, CT
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithnm and an empirical binding free energy function. J Comput Chem 19:1639–1662
Morris GM, Goodsell DS, Huey R, Olson AJ (1996) Distributed automated docking of flexible ligands to proteins parallel applications of AutoDock 2.4. J Comput Aided Mol Des 10:293–304
Case DA, Cheatham TA, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvary I, Wong KF, Paesani F, Vanicek J, Wu X, Bronzell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA (2008) AMBER 10. University of California, San Francisco, CA
Bayly CI, Cieplak P, Cornell WD, Kollman PA (1993) A well-behaved electrostatic potential based method using charge restraints for determining atom-centered charges: the RESP model. J Phys Chem 97:10269–11028
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc 117:5179–5197
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general Amber force field. J Comput Chem 25:1157–1174
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341
Darden T, York D, Pedersen L (1998) Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Srinivasan J, Cheatham TE, Cieplak P, Kollman PA, Case DA (1998) Continuum solvent studies of the stability of DNA, RNA, and Phosphoramidate-DNA Helices. J Am Chem Soc 120:9401–9409
Saíz-Urra L, Cabrera MA, Froeyen M (2011) Exploring the conformational changes of the ATP binding site of gyrase B from Escherichia colicomplexed with different established inhibitors by using molecular dynamics simulation: Protein – ligand interactions in the light of the alanine scanning and free energy decomposition methods. J Mol Graph Model 29:726–739
El-Barghouthi MI, Jaime C, Al-Sakhen NA, Issa AA, Abdoh AA, Omari MM, Badwan AA, Zughul MB (2008) Molecular dynamics simulations and MM—PBSA calculations of the cyclodextrin inclusion complexes with 1-alkanols, para-substituted phenols and substituted imidazoles. J Mol Struct THEOCHEM 853:45–52
Cong XJ, Tan JJ, Liu M, Chen W, Wang CX (2010) Computational study of binding mode for N-substituted pyrrole derivatives to HIV-1 gp41. Prog Biochem Biophys 37(8):904–91566
Andricioaei I, Karplus M (2001) On the calculation of entropy from covariance matrices of the atomic fluctuations. J Chem Phys 115:6289–6292
Lobanov MY, Bogatyreva NS, Galzitskaya OV (2008) Radius of gyration as an indicator of protein structure compactness. Mol Biol 42:623–628
Vivier I, Marguet D, Naquet P, Bonicel J, Black D, Li CXY, Bernard AM, Gorvel JP, Pierres M (1991) Evidence that thymocyte-activating molecule is mouse cd26 (dipeptidyl peptidase-iv). J Immunol 147:447–454
Tanaka S, Murakami T, Horikawa H, Sugiura M, Kawashima K, Sugita T (1997) Suppression of arthritis by the inhibitors of dipeptidyl peptidase IV Int. J Immunopharmacol 19:15–24
Bristol LA, Sakaguchi K, Appella E, Doyle D, Taka’cs L (1992) Thymocyte costimulating antigen is CD26 (Dipeptidyl peptidase-IV)–costimulation of granulocyte, macrophage, and T-lineageceli-proliferation via CD26. J Immunol 149:367–372
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPOLT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8:127–134
Hu GD, Zhu T, Zhang SL, Wang D, Zhang QG (2010) Some insights into mechanism for binding and drug resistance of wild type and I50V V82A and I84V mutations in HIV-1 protease with GRL-98065 inhibitor from molecular dynamic simulations. Eur J Med Chem 45:227–235
Wu EL, Han K, Zhang JZ (2008) Selectivity of neutral/weakly basic P1 group inhibitors of thrombin and trypsin by a molecular dynamics study. Chem Eur J 14:8704–8714
Acknowledgments
This work is supported by grants from the National Science Foundation of China (Nos. 21276122, 21136001, and 20876073) and State Key Laboratory of Materials-Oriented Chemical Engineering, College of Chemistry and Chemical Engineering, Nanjing University of Technology of China (No. ZK201212).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gu, Y., Wang, W., Zhu, X. et al. Molecular dynamic simulations reveal the mechanism of binding between xanthine inhibitors and DPP-4. J Mol Model 20, 2075 (2014). https://doi.org/10.1007/s00894-014-2075-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-014-2075-1