Abstract
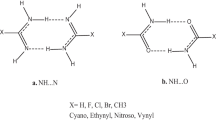
We investigate the changes in the solvation properties of the natural nucleic acid bases due to the formation of the canonical Watson–Crick hydrogen-bonded complexes. To this end, the changes in the free energy of solvation of the bases induced upon hydrogen-bonded dimerization are analyzed by means of the hydrophobic similarity index, which relies on the atomic contributions to the free energy of solvation determined by the partitioning method implemented in the framework of the MST continuum model. Such an index is also used to examine the hydrophobic similarity between the canonical nucleic acid bases and a series of highly apolar analogues, which have been designed as potential candidates to expand the genetic alphabet. The ability of these analogues to be incorporated into modified DNA duplexes can be related to the large reduction in the hydrophilicity of the natural bases upon formation of the canonical hydrogen-bonded dimers. The results illustrate the suitability of the hydrophobic similarity index to rationalize the role played by solvation in molecular recognition.



Similar content being viewed by others
References
Bleicher KH, Bohm HJ, Muller K, Alanine AI (2003) Nat Rev Drug Discov 52:369–378
Schuffenhauer A, Popov M, Schopfer U, Acklin P, Stanek J, Jacoby E (2004) Comb Chem High Throughput Screen 7:771–781
Dean PM (ed) (1995) Molecular similarity in drug design. Blackie Academic, London
Johnson MA, Maggiora GM (eds) (1990) Concepts and applications of molecular similarity. Wiley, New York
Carbó-Dorca R, Gironés X, Mezey PG (eds) (2001) Fundamentals of molecular similarity. Kluwer Academic/Plenum Publishers, New York
Carbó R, Leyda L, Arnau M (1980) Int J Quant Chem 17:1185–1189
Carbó-Dorca R, Robert D, Amat L, Girones X, Besalú E (2000) Lecture notes in chemistry, vol 73. Springer, Berlin, Hiedelberg, New York
Lee C, Smithline S (1994) J Phys Chem 98:1135–1138
Bowen-Jenkins PE, Richards WG (1986) J Chem Soc, Chem Commun 133–135
Popelier PLA (1999) J Phys Chem A 103:2883–2890
O’Brien SE, Popelier PLA (1999) Can J Chem 77:28–36
Kier LB, Hall LH (1986) Molecular connectivity letchworkin structure-activity analysis. Research Studies, Letchwork
Hall LH, Kier LB (1991) The molecular connectivity chi indexes and kappa shape indexes in structure-property relations. In: Lipkowitz KB, Boyd DB (eds) Reviews in Computational Chemistry, vol 2. VCH, New York pp 367–442
Luque FJ, Sanz F, Illas F, Pouplana R, Smeyers YG (1988) Eur J Med Chem 23:7–10
Richard AM (1991) J Comput Chem 12:959–969
Hernández B, Orozco M, Luque FJ (1996) J Comput Aided Mol Des 10:535–544
Náray-Szabó G, Ferenczy GG (1995) Chem Rev 95:829–847
Hodgkin EE, Richards WG (1987) Int J Quant Chem 14:105–110
Burt C, Richards WG, Huxley P (1990) J Comput Chem 11:1139–1146
Manaut F, Sanz F, José J, Milesi M (1991) J Comput Aided Mol Des 5:371–380
Petke JD (1993) J Comput Chem 8:928–933
Rodríguez J, Manaut F, Sanz F (1993) J Comput Chem 14:922–927
Thorner DA, Willet P, Wright PM, Taylor R (1997) J Comput Aided Mol Des 11:163–174
Meyer AM, Richards WG (1991) J Comput Aided Mol Des 5:427–439
Mezey PG (1993) Shape in chemistry: an introduction to molecular shape and topology. VCH, New York
Tokarski JS, Hopfinger AJ (1994) J Med Chem 37:3639–3654
Jain N, Dietterich TG, Lathrop RH, Chapman D, Critchlow Jr RE, Bauer BE, Webster TA, Lozano-Pérez T (1994) J Comput Aided Mol Des 8:635–652
Cramer RD III, Paterson DE, Bunce JD (1988) J Am Chem Soc 110:5959–5967
Klebe G, Mietzner T, Weber F (1994) J Comput Aided Mol Des 8:751–778
Perkins TDJ, Mills JEJ, Dean PM (1995) J Comput Aided Mol Des 9:479–490
Mestres J, Rohrer DC, Maggiora GM (1997) J Comput Chem 18:934–954
Kubinyi H (ed) 3D QSAR in drug design: theory, methods and applications. (1993) ESCOM, Leiden
Lemmen C, Lengauer T, Klebe G (1998) J Med Chem 41:4502–4520
Miller MD, Sheridan RP, Kearsley K (1999) J Med Chem 42:1505–1514
Palm K, Luthman K, Ungell AL, Strandlund G, Artursson P (1996) J Pharm Sci 85:32–39
Palm K, Luthman K, Ungell AL, Strandlund G, Beigi F, Lundahl P, Artursson P (1998) J Med Chem 41:5382–5392
Clark DE (1999) J Pharm Sci 88:807–814
Clark DE (1999) J Pharm Sci 88:815–821
Kantola A, Villar HO, Loew GH (1991) J Comput Chem 12:681–689
Segarra V, López M, Ryder H, Palacios JM (1999) Quant Struct- Act Relatsh 18:474–481
Sulea T, Purísima EO (1999) Quant Struct- Act Relatsh 18:154–158
Eisenberg D, McLachlan AD (1986) Nature 319:199–203
Eisenberg D, Schwarz E, Komaromy M, Wall R (1984) J Mol Biol 179:125–142
Barril X, Muñoz J, Luque FJ, Orozco M (2000) Phys Chem Chem Phys 2:4897–4905
Muñoz J, Barril X, Luque FJ, Gelpí JL, Orozco M (2001) Partitioning of free energies of solvation into fragment contributions: applications in drug design. In: Carbó-Dorca R, Gironés X, Mezey PG (eds) Fundamentals of molecular similarity. Kluwer Academic Plenum Publishers, New York pp 143–168
Audry E, Dubost JP, Colleter JC, Dallet P (1986) Eur J Med Chem 21:71–72
Brasseur R (1991) J Biol Chem 266:16120–16127
Heiden FW, Moeckel G, Brickmann J (1993) J Comput Aided Mol Des 7:503–514
Kellogg E, Semus SF, Abraham DJ (1991) J Comput Aided Mol Des 5:545–552
Gaillard P, Carrupt PA, Testa B, Boudon A (1994) J Comput Aided Mol Des 8:83–96
Furet P, Sele A, Cohén, NCJ (1988) J Mol Graph 6:182–189
Croizet F, Langlois MH, Dubost JP, Braquet P, Audry E, Dallet P, Colleter JC (1990) J Mol Graph 8:153–155
Fauchére JL, Quarendon P, Kaetterer L (1988) J Mol Graph 6:203–206
Du Q, Arteca GA (1996) J Comput Aided Mol Des 10:133–144
Du Q, Arteca GA, Mezey PG (1997) J Comput Aided Mol Des 11:503–516
Bone RGA, Villar HO (1995) J Mol Graph 13:201–208
Muñoz J, Hernández B, Barril X, Orozco M, Luque FJ (2002) J Comput Chem 23:554–563
Luque FJ, Barril X, Orozco M (1999) J Comput Aided Mol Des 13:139–152
Curutchet C, Orozco M, Luque FJ (2001) J Comput Chem 22:1180–1193
Luque FJ, Curutchet C, Muñoz-Muriedas J, Bidon-Chanal A, Morreale A, Gelpi JL, Orozco M (2003) Phys Chem Chem Phys 5:3827–3836
Miertus S, Scrocco E, Tomasi J (1981) Chem Phys 55:117–129
Pierotti RA (1976) Chem Rev 76:717–726
Claverie P (1978) In: Pullman B (ed) Intermolecular interactions: from diatomics to biomolecules. Wiley, Chichester
Luque FJ, Bofill JM, Orozco M (1995) J Chem Phys 103:10183–10191
Orozco M, Bachs M, Luque FJ (1995) J Comput Chem 16:563–575
Peterson M, Poirier, R MonsterGauss. Department of Biochemistry, Univ Toronto, Canada. Version modified by Cammi R, Tomasi J (1987) and by Curutchet C, Orozco M, Luque FJ (2004)
Huertas O, Orozco M, Luque FJ (2006) J Phys Chem A 110:510–518
Hobza P, Sponer J (1999) Chem Rev 99:3247–3276
Bader RFW (1991) Chem Rev 91:893–928
Silvi B, Savin A (1994) Nature 31:683–686
Alkorta I, Rozas I, Elguero J (1998) Struct Chem 9:243–248
Muñoz J, Sponer J, Hobza P, Orozco M, Luque FJ (2001) J Phys Chem B 105:6051–6060
Fuster F, Silvi B (2000) Theor Chem Acc 104:13–21
Orozco M, Cubero E, Barril X, Colominas C, Luque FJ (1999) Nucleic acid bases in solution. In: Leszczynski J (ed) Computational molecular biology. Theoretical computational chemistry, vol 8 Elsevier, Amsterdam, pp 119–165
Wu Y, Ogawa AK, Berger M, McMinn DL, Schuitz PG, Romesberg FE (2000) J Am Chem Soc 122:7621–7632
Loakes D (2001) Nucleic Acids Res 29:2437–2447
Hansch C, Leo A (1995) Exploring QSAR: hydrophobic, electronic and steric constants. American Chemical Society, Washington
Acknowledgments
We thank the Ministerio de Ciencia y Tecnología (grants CTQ2005-09365 and BIO2003-06848) for financial assistance and the Centre de Supercomputació de Catalunya for computational facilities.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Proceedings of “Modeling Interactions in Biomolecules II”, Prague, September 5th–9th, 2005.
Rights and permissions
About this article
Cite this article
Muñoz-Muriedas, J., Barril, X., López, J.M. et al. A hydrophobic similarity analysis of solvation effects on nucleic acid bases. J Mol Model 13, 357–365 (2007). https://doi.org/10.1007/s00894-006-0150-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-006-0150-y