Abstract
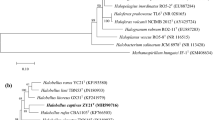
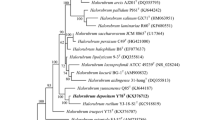
Two halophilic archaeal strains, R30T and tADLT, were isolated from an aquaculture farm in Dailing, China, and from Deep Lake, Antarctica, respectively. Both have rod-shaped cells that lyse in distilled water, stain Gram-negative and form red-pigmented colonies. They are neutrophilic, require >120 g/l NaCl and 48–67 g/l MgCl2 for growth but differ in their optimum growth temperatures (30 °C, tADLT vs. 40 °C, R30T). The major polar lipids were typical for members of the Archaea but also included a major glycolipid chromatographically identical to sulfated mannosyl glucosyl diether (S-DGD-1). The 16S rRNA gene sequences of the two strains are 97.4 % identical, show most similarity to genes of the family Halobacteriaceae, and cluster together as a distinct clade in phylogenetic tree reconstructions. The rpoB′ gene similarity between strains R30T and tADLT is 92.9 % and less to other halobacteria. Their DNA G + C contents are 62.4–62.9 mol % but DNA–DNA hybridization gives a relatedness of only 44 %. Based on phenotypic, chemotaxonomic and phylogenetic properties, we describe two new species of a novel genus, represented by strain R30T (= CGMCC 1.10593T = JCM 17270T) and strain tADLT (= JCM 15066T = DSMZ 22187T) for which we propose the names Halohasta litorea gen. nov., sp. nov. and Halohasta litchfieldiae sp. nov., respectively.

Similar content being viewed by others
References
Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML (2004) Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl Environ Microbiol 70:5258–5265
Burns DG, Janssen PH, Itoh T, Kamekura M, Echigo A, Dyall-Smith ML (2010) Halonotius pteroides gen. nov., sp. nov., an extremely halophilic archaeon recovered from a saltern crystallizer in southern Australia. Int J Syst Evol Microbiol 60:1196–1199
Cui HL, Lin ZY, Dong Y, Zhou PJ, Liu SJ (2007) Halorubrum litoreum sp. nov., an extremely halophilic archaeon from a solar saltern. Int J Syst Evol Microbiol 57:2204–2206
Cui HL, Zhou PJ, Oren A, Liu SJ (2009) Intraspecific polymorphism of 16S rRNA genes in two halophilic archaeal genera, Haloarcula and Halomicrobium. Extremophiles 13:31–37
Cui HL, Gao X, Sun FF, Dong Y, Xu XW, Zhou YG, Liu HC, Oren A, Zhou PJ (2010a) Halogranum rubrum gen. nov., sp. nov., a halophilic archaeon isolated from a marine solar saltern. Int J Syst Evol Microbiol 60:1366–1371
Cui HL, Gao X, Yang X, Xu XW (2010b) Halorussus rarus gen. nov., sp. nov., a new member of the family Halobacteriaceae isolated from a marine solar saltern. Extremophiles 14:493–499
Cui HL, Gao X, Yang X, Xu XW (2011a) Halolamina pelagica gen. nov., sp. nov., a new member of the family Halobacteriaceae. Int J Syst Evol Microbiol 61:1617–1621
Cui HL, Yang X, Mou YZ (2011b) Salinarchaeum laminariae gen. nov., sp. nov.: a new member of the family Halobacteriaceae isolated from salted brown alga Laminaria. Extremophiles 15:625–631
Cui HL, Mou YZ, Yang X, Zhou YG, Liu HC, Zhou PJ (2012) Halorubellus salinus gen. nov., sp. nov. and Halorubellus litoreus sp. nov., novel halophilic archaea isolated from a marine solar saltern. Syst Appl Microbiol 35:30–34
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Dussault HP (1955) An improved technique for staining red halophilic bacteria. J Bacteriol 70:484–485
Dyall-Smith ML (2009) The Halohandbook: protocols for haloarchaeal genetics. http://www.haloarchaea.com/resources/halohandbook
Gonzalez C, Gutierrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov. an amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24:710–715
Gutiérrez C, González C (1972) Method for simultaneous detection of proteinase and esterase activities in extremely halophilic bacteria. Appl Microbiol 24:516–517
Gutiérrez MC, Castillo AM, Kamekura M, Ventosa A (2008) Haloterrigena salina sp. nov., an extremely halophilic archaeon isolated from a salt lake. Int J Syst Evol Microbiol 58:2880–2884
Huß VAR, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192
Kamekura M, Mizuki T, Usami R, Yoshida Y, Horikoshi K, Vreeland RH (2004) The potential use of signature bases from 16S rRNA gene sequences to aid the assignment of microbial strains to genera of halobacteria. In: Ventosa A (ed) Halophilic microorganisms. Springer, Heidelberg, pp 77–87
Makhdoumi-Kakhki A, Amoozegar MA, Bagheri M, Ramezani M, Ventosa A (2012a) Haloarchaeobius iranensis gen. nov., sp. nov., an extremely halophilic archaeon isolated from the saline lake Aran-Bidgol. Iran Int J Syst Evol Microbiol 62:1021–1026
Makhdoumi-Kakhki A, Amoozegar MA, Ventosa A (2012b) Halovenus aranensis gen. nov., sp. nov., a novel extremely halophilic archaeon from Aran-Bidgol salt lake. Iran Int J Syst Evol Microbiol 62:1331–1336
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
McDade JJ, Weaver RH (1959) Rapid methods for the detection of gelatin hydrolysis. J Bacteriol 77:60–64
Minegishi H, Kamekura M, Itoh T, Echigo A, Usami R, Hashimoto T (2010) Further refinement of Halobacteriaceae phylogeny based on the full-length RNA polymerase subunit B’ (rpoB ’) gene. Int J Syst Evol Microbiol 60:2398–2408
Oren A (2006) The order Halobacteriales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The prokaryotes, vol 3, vol 3, 3rd edn. Springer, New York, pp 113–164
Oren A, Ventosa A, Grant WD (1997) Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Bacteriol 47:233–238
Owen RJ, Pitcher D (1985) Current methods for estimating DNA base composition and levels of DNA–DNA hybridization. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 67–93
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 30970006), the opening project of State key Laboratory of Microbial Resources (No. SKLMR-20100604, Institute of Microbiology, Chinese Academy of Sciences) and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). Deep Lake water samples were kindly supplied by J. Hoffman (J. Craig Venter Inst., USA) and R. Cavicchioli (University of NSW, Sydney, Australia). M. D.-S. is grateful to D. Oesterhelt (Max-Planck Institute, Munich, Germany) for providing laboratory resources for part of this study. We thank Jean Euzéby (École National Vétérinaire, Toulouse, France) for his suggestion and advice regarding the formulation of genus and species names.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by A. Oren.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mou, YZ., Qiu, XX., Zhao, ML. et al. Halohasta litorea gen. nov. sp. nov., and Halohasta litchfieldiae sp. nov., isolated from the Daliang aquaculture farm, China and from Deep Lake, Antarctica, respectively. Extremophiles 16, 895–901 (2012). https://doi.org/10.1007/s00792-012-0485-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-012-0485-5