Abstract
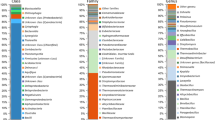
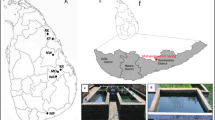
Prokaryotic diversities of 12 geothermal hot springs located in Northern, Central and Southern Tunisia were investigated by culture-based and molecular approaches. Enrichment cultures for both aerobic and anaerobic microorganisms were successfully obtained at temperatures ranging from 50 to 75°C. Fourteen strains including four novel species were cultivated and assigned to the phyla Firmicutes (9), Thermotogae (2), Betaproteobacteria (1), Synergistetes (1) and Bacteroidetes (1). Archaeal or universal oligonucleotide primer sets were used to generate 16S rRNA gene libraries. Representative groups included Proteobacteria, Firmicutes, Deinococcus-Thermus, Thermotogae, Synergistetes, Bacteroidetes, Aquificae, Chloroflexi, candidate division OP9 in addition to other yet unclassified strains. The archaeal library showed a low diversity of clone sequences belonging to the phyla Euryarchaeota and Crenarchaeota. Furthermore, we confirmed the occurrence of sulfate reducers and methanogens by amplification and sequencing of dissimilatory sulfite reductase (dsrAB) and methyl coenzyme M reductase α-subunit (mcrA) genes. Altogether, we discuss the diverse prokaryotic communities arising from the 12 geothermal hot springs studied and relate these findings to the physico-chemical features of the hot springs.






Similar content being viewed by others
References
Alfredsson GA, Kristjansson JK, Hjorleifsdottir S, Stetter KO (1988) Rhodothermus marinus, gen. nov., sp. nov., a thermophilic, halophilic bacterium from submarine hot springs in Iceland. J Gen Microbiol 134:299–306
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Antunes A, Taborda M, Huber R, Moissl C, Nobre MF, da Costa MS (2008) Halorhabdus tiamatea sp. nov., a non-pigmented, extremely halophilic archaeon from a deep-sea, hypersaline anoxic basin of the Red Sea, and emended description of the genus Halorhabdus. Int J Syst Evol Microbiol 58:215–220
Arnon S, Ronen Z, Adar E, Yakirevich A, Nativ R (2005) Two-dimensional distribution of microbial activity and flow patterns within naturally fractured chalk. J Contam Hydrol 19:165–186
Baross JA (1995) Isolation growth and maintenance of hyperthermophiles. In: Robb FT, Place AR (eds) Archaea a laboratory manual, thermophiles. Cold Spring Harbor Laboratory, New York, pp 15–23
Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523
Bouri S, Makni J, Ben Dhia H (2008) A synthetic approach integrating surface and subsurface data for prospecting deep aquifers: the southeast Tunisia. Environ Geol 54:1473–1484
Bredholt S, Jacob Sonne-Hansen J, Nielsen P, Mathrani IM, Ahring BK (1999) Caldicellulosiruptor kristjanssonii sp. nov., a cellulolytic extremely thermophilic anaerobic bacterium. Int J Syst Bacteriol 49:991–996
Casamayor EO, Pedrós-Alió C, Muyzer G, Amann R (2002) Microheterogeneity in 16S ribosomal DNA-defined bacterial populations from a stratified planktonic environment is related to temporal changes and to ecological adaptations. Appl Environ Microbiol 68:1706–1714
Cavicchioli R (2002) Extremophiles and the search for extraterrestrial life. Astrobiology 2:281–292
Chin KJ, Hahn D, Hengstmann U, Liesack W, Janssen PH (1999) Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Appl Environ Microbiol 65:5042–5049
Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM (2003) The ribosomal database project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res 1:442–443
Curtis TP, Sloan WT (2005) Exploring microbial diversity. Science 309:1331–1333
Dayhoff MO, Schwartz RM, Orcutt BC (1978) A model for evolutionary change in proteins. In: Dayhoff MO (ed) Atlas of protein sequence and structure. Natl Biomed Res Foundation, Washington, DC, pp 345–352
Davidova IE, Duncan KE, Choi OK, Suflita JM (2006) Desulfoglaeba alkanexedens, gen. nov., sp. nov., an n-alkane-degrading sulfate-reducing bacterium. Int J Syst Evol Microbiol 56:2737–2742
Degryse E, Glansdorff N, Pierard A (1978) A comparative analysis of extreme thermophilic bacteria belonging to the genus Thermus. Arch Microbiol 117:189–196
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci USA 89:5685–5689
Demirjian DC, Moris-Varas F, Cassidy CS (2001) Enzymes from extremophiles. Curr Opin Chem Biol 5:144–151
Eden P, Schmidt T, Blakemore R, Pace N (1991) Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction amplified 16S ribosomal RNA specific DNA. Int J Syst Bacteriol 41:324–325
Elshahed MS, Najar FZ, Roe BA, Oren A, Dewers TA, Krumholz LR (2004) Survey of archaeal diversity reveals an abundance of halophilic Archaea in a low-salt sulfide- and sulfur-rich spring. Appl Environ Microbiol 70:2230–2239
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103:626–631
Godon JJ, Zumstein E, Dabert P, Habouzit F, Moletta R (1997) Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequences analysis. Appl Environ Microbiol 63:2802–2813
Hales BA, Edwards C, Ritchie DA, Hall G, Pickup RW, Saunders JR (1996) Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequences analysis. Appl Environ Microbiol 62:668–675
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Haouari O, Fardeau ML, Cayol JL, Fauque G, Casiot C, Elbaz-Poulichet F, Hamdi M, Ollivier B (2008) Thermodesulfovibrio hydrogeniphilus sp. nov., a new thermophilic sulphate-reducing bacterium isolated from a Tunisian hot spring. Syst Appl Microbiol 31:38–42
Hengstmann U, Chin KJ, Janssen PH, Liesack W (1999) Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numerically abundant culturable bacteria from an anoxic rice paddy soil. Appl Environ Microbiol 65:5050–5058
Hetzer A, Morgan HW, McDonald IR, Daughney CJ (2007) Microbial life in Champagne Pool, a geothermal spring in Waiotapu, New Zealand. Extremophiles 11:605–614
Hugenholtz P, Hooper SD, Kyrpides NC (2009) Genomics update focus: Synergistetes. Environ Microbiol 11:1327–1329
Jellouli A (2002) La Tunisie Thermale. Deuxième édition, ministère du tourisme des loisirs et de l’artisanat, Tunis, ISBN: 9973-815-39-4
Jiang H, Dong H, Yu B, Liu X, Li Y, Ji S, Zhang CL (2007) Microbial response to salinity change in Lake Chaka, a hypersaline lake on Tibetan plateau. Environ Microbiol 9:2603–2621
Kanokratana P, Chanapan S, Pootanakit K, Eurwilaichitr L (2004) Diversity and abundance of bacteria and archaea in the Bor Khlueng hot spring in Thailand. J Basic Microbiol 44:430–444
Klein M, Friedrich M, Roger AJ, Hugenholtz P, Fishbain S, Abicht H, Blackall LL, Stahl DA, Wagner A (2001) Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J Bacteriol 183:6028–6035
Lai Q, Yuan J, Wu C, Shao Z (2009) Oceanibaculum indicum gen. nov., sp. nov., isolated from deep seawater of the Indian ocean. Int J Syst Evol Microbiol 59:1733–1737
Lamberti GA, Resh VH (1983) Geothermal effects on stream benthos: separate influences of thermal and chemical components on periphyton and macroinvertebrates. Can J Fish Aquat Sci 40:1995–2009
Lösekann T, Knittel K, Nadalig T, Fuchs B, Niemann H, Boetius A, Amann R (2007) Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby mud volcano, Barents sea. Appl Environ Microbiol 73:3348–3362
Ludwig W, Strunk O, Klugbauer S, Weizenegger M, Neumaier J, Bachleitner M, Schleifer KH (1998) Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554–568
Lueders T, Chin KJ, Conrad R, Friedrich M (2001) Molecular analyses of methyl-coenzyme M reductase α-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ Microbiol 3:194–204
Madigan MT, Martinko JM (2006) Brock biology of microorganisms, 11th edn. Pearson Education International, London
Magot M (2005) Indigenous microbial communities in oil fields. In: Ollivier B, Magot M (eds) Petroleum microbiology. ASM Press, Washington, pp 21–34
Marchesi JR, Sato T, Weightman A, Martin TA, Fry JC, Hiom SJ, Wade WG (1998) Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64:795–799
Mori K, Suzuki KI (2008) Thiofaba tepidiphila gen. nov., sp. nov., a novel obligately chemolithoautotrophic sulfur-oxidizing bacterium of the Gammaproteobacteria isolated from a hot spring. Int J Syst Evol Microbiol 58:1885–1891
Muyzer G, Stams AJM (2008) The ecology and biotechnology of sulfate-reducing bacteria. Nat Rev Microbiol 6:441–454
Namwong S, Hiraga K, Takada K, Tsunemi M, Tanasupawat S, Oda K (2006) A halophilic serine proteinase from Halobacillus sp SR5-3 isolated from fish sauce: purification and characterization. Biosci Biotechnol Biochem 70:1395–1401
Newman DK, Kennedy EK, Coates JD, Ahmann D, Ellis DJ, Lovley DR, Morel FMM (1997) Dissimilatory arsenate and sulfate reduction in Desulfotomaculum auripigmentum sp. nov. Arch Microbiol 168:380–388
Nold SC, Zwart G (1998) Patterns and governing forces in aquatic microbial communities. Aquat Ecol 32:17–35
Nunoura T, Hirayama H, Takami H, Oida H, Nishi S, Shimamura S, Suzuki Y, Inagaki F, Takai K, Nealson KH, Horikoshi K (2005) Genetic and functional properties of uncultivated thermophilic crenarchaeotes from a subsurface gold mine as revealed by analysis of genome fragments. Environ Microbiol 7:1967–1984
Pentecost A (1996) High temperature ecosystems and their chemical interactions with their environment. In: Brock GR, Groode JA (eds) Ciba foundation symposium, evolution of hydrothermal ecosystems on earth (and Mars?). Ciba Fdn, London, pp 99–111
Pham VD, Hnatow LL, Zhang S, Fallon RD, Jackson SC, Tomb JF, DeLong EF, Keeler SJ (2009) Characterizing microbial diversity in production water from an Alaskan mesothermic petroleum reservoir with two independent molecular methods. Environ Microbiol 11:176–187
Purcell D, Sompong U, Yim LC, Barraclough TG, Peerapornpisal Y, Pointing S (2006) The effects of temperature pH and sulphide on the community structure of hyperthermophilic streamers in hot springs of northern Thailand. FEMS Microbiol Ecol 60:456–466
Purdy KJ, Cresswell-Maynard TD, Nedwell DB, McGenity TJ, Grant WD, Timmis KN, Embley TM (2004) Isolation of haloarchaea that grow at low salinities. Environ Microbiol 6:591–595
Ravot G, Magot M, Fardeau ML, Patel B, Prensier G, Egan A, Garcia JL, Ollivier B (1995) Thermotoga elfii sp. nov., a novel thermophilic bacterium from an African oil-producing well. Int J Syst Bacteriol 45:314–318
Roh H, Yu CP, Fuller ME, Chu KH (2009) Identification of hexahydro-1,3,5-trinitro-1,3,5-triazine-degrading microorganisms via 15N-stable isotope probing. Environ Sci Technol 43:2505–2511
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schiraldi C, De Rosa M (2002) The production of biocatalysts and biomolecules from extremophiles. Trends Biotechnol 20:515–521
Schleheck D, Tindall BJ, Rosselló-Mora R, Cook AM (2004) Parvibaculum lavamentivorans gen. nov., sp. nov., a novel heterotroph that initiates catabolism of linear alkylbenzenesulfonate. Int J Syst Evol Microbiol 54:1489–1497
Siboni N, Ben-Dov E, Sivan A, Kushmaro A (2008) Global distribution and diversity of coral-associated Archaea and their possible role in the coral holobiont nitrogen cycle. Environ Microbiol 10:2979–2990
Soo RM, Wood SA, Grzymski JJ, McDonald IR, Cary SC (2009) Microbial biodiversity of thermophilic communities in hot mineral soils of Tramway Ridge Mount Erebus Antarctica. Environ Microbiol 11:715–728
Spear JR, Walker JJ, Mc Collom TM, Pace N (2005) Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc Natl Acad Sci USA 7:2555–2560
Springer E, Sachs MS, Woese CR, Boone DR (1995) Partial gene-sequences for the α-subunit of methyl-coenzyme M reductase (MCRI) as a phylogenetic tool for the family Methanosarcinaceae. Int J Syst Bacteriol 45:554–559
Stackebrandt E, Ebers J (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:153–155
Stein LY, Jones G, Alexander B, Elmund K, Wright-Jones C, Nealson KH (2002) Intriguing microbial diversity associated with metal-rich particles from a freshwater reservoir. FEMS Microbiol Ecol 42:431–440
Suzuki D, Ueki A, Amaishit A, Ueki K (2007) Desulfopila aestuarii gen. nov., sp. nov., a Gram-negative rod-like sulfate-reducing bacterium isolated from an estuarine sediment in Japan. Int J Syst Evol Microbiol 57:520–526
Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK (2001) Archaeal diversity in waters from deep South African gold mines. Appl Environ Microbiol 67:5750–5760
Teske AK, Hinrichs U, Edgcomb V, Gomez AV, Kysela D, Sylva SP, Sogin ML, Jannasch HW (2002) Microbial diversity of hydrothermal sediments in the Guaymas Basin evidence for anaerobic methanotrophic communities. Appl Environ Microbiol 68:1994–2007
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Walsh DAR, Papke RT, Doolittle WF (2005) Archaeal diversity along a soil salinity gradient prone to disturbance. Environ Microbiol 7:1655–1666
Widdel F, Pfennig N (1981) Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. Isolation of new sulfate-reducing bacteria enriched with acetate from saline environments. Description of Desulfobacter postgatei gen. nov., sp. nov. Arch Microbiol 129:395–400
Zouari K, Mamou A (1992) Les systèmes aquifères du Sud tunisien caractéristiques hydrochimiques et isotopiques. Conditions de recharge et optimisation de la gestion. Revue de la Faculté des sciences de Marrakech, Semlalia, pp 41–44
Acknowledgments
We are grateful to Dr. E. Roussel and the director of the Tunisian Office of Balneology for their help. We also acknowledge Prof. J. P. Euzéby for support in the Latin etymologies of species names. This research was financed by the Ministry of Higher Education and Scientific Research in Tunisia and by CNRS (Centre National de la Recherche Scientifique).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Oren.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sayeh, R., Birrien, J.L., Alain, K. et al. Microbial diversity in Tunisian geothermal springs as detected by molecular and culture-based approaches. Extremophiles 14, 501–514 (2010). https://doi.org/10.1007/s00792-010-0327-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-010-0327-2