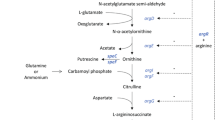
Abstract
Already very early, the study of microbial arginine biosynthesis and its regulation contributed significantly to the development of new ideas and concepts. Hence, the term “repression” was proposed by Vogel (The chemical basis of heredity, The John Hopkins Press, Baltimore, 1957) (in opposition to induction) to describe the relative decrease in acetylornithinase production in Escherichia coli cells upon arginine supplementation, whereas the term “regulon” was coined by Maas and Clark (J Mol Biol 8:365–370, 1964) for the ensemble of arginine biosynthetic genes dispersed over the E. coli chromosome but all subjected to regulation by the trans-acting argR gene product. Since then, unraveling of the molecular mechanisms controlling arginine biosynthesis, catabolism, and transport in and out the cell, have revealed moonlighting activities of enzymes and transcriptional regulators that generate unexpected interconnections between at first sight totally unrelated cellular processes, and have continued to replenish scientific knowledge and stimulated creative thinking. Furthermore, arginine is much more than just a common amino acid for protein synthesis. It may also be used as sole source of nitrogen by E. coli and a source of nitrogen, carbon and energy by many other bacteria. It is a substrate for the synthesis of polyamines, and important for the extreme acid resistance of E. coli. Furthermore, the guanidino group of arginine is well suited to engage in multiple interactions involving hydrogen bonds and ionic interactions with proteins and nucleic acids. Here, we combine major historical discoveries with current state of the art knowledge on arginine biosynthesis, catabolism and transport, and especially the regulation of these processes in E. coli, with reference to other microorganisms.






Similar content being viewed by others
References
Abadjieva A, Hilven P, Pauwels K, Crabeel M (2000) The yeast ARG7 gene product is autoproteolyzed to two subunit peptides, yielding active ornithine acetyltransferase. J Biol Chem 275:11361–11367
Abadjieva A, Pauwels K, Hilven P, Crabeel M (2001) A new yeast metabolon involving at least the first two enzymes of arginine biosynthesis: acetylglutamate synthase activity requires complex formation with acetylglutamate kinase. J Biol Chem 276:42869–42880
Abd-El-Al A, Ingraham J (1969) Control of carbamoylphosphate synthesis in Salmonella typhimurium. J Biol Chem 244:4033–4038
Abdelal AT, Nainan OV (1979) Regulation of N-acetylglutamate synthesis in Salmonella typhimurium. J Bacteriol 137:1040–1042
Aleshin VV, Zakataeva NP, Livshits VA (1999) A new family of amino-acid-efflux proteins. Trends Biochem Sci 24:133–135
Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A (1999) Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol 181:6361–6370
Ames GFL, Nikaido K, Wang IX, Liu PQ, Liu CE, Hu C (2001) Purification and characterization of the membrane-bound complex of an ABC transporter, the histidine permease. J Bioenerget Biomembr 33:79–92
Andrell J, Hicks MG, Palmer T, Carpenter EP, Iwata S, Maher MJ (2009) Crystal structure of the acid-inducible arginine decarboxylase from Escherichia coli: reversible decamer assembly controls enzyme activity. Biochemistry 48:3915–3927
Asi AM, Rahman NA, Merican AF (2003) Applications of the linear interaction energy method (LIE) to estimate the binding free energy values of Escherichia coli wild-type and mutant arginine repressor C-terminal domain (ArgRc)-l-arginine and ArgRc-l-citrulline protein-ligand complexes. J Mol Graph Model 22:249–262
Auger EA, Redding KE, Plumb T, Childs LC, Meng SY, Bennett GN (1989) Construction of lac fusions to the inducible arginine and lysine decarboxylase genes of Escherichia coli K-12. Mol Microbiol 3:609–620
Azam TA, Ishihama A (1999) Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J Biol Chem 274:33105–33113
Bacon DF, Vogel HJ (1963) A regulatory gene simultaneously involved in repression and induction. Cold Spring Harbor Symp Quant Biol 28:437–438
Baumberg S (1970) Acetylhistidine as substrate for acetylornithinase: a new system for the selection of arginine regulation mutants in Escherichia coli. Mol Gen Genet 106:162–173
Bellman A, Vrljic M, Patek M, Krämer R, Eggeling L (2001) Expression control and specificity of the basic amino acid transporter LysE of Corynebacterium glutamicum. Microbiology 147:1765–1774
Bény G, Cunin R, Glansdorff N, Boyen A, Charlier J, Kelker N (1982) Transcription of regions within the divergent argECBH operon of Escherichia coli: evidence for lack of an attenuation mechanism. J Bacteriol 151:58–61
Bi H, Zhang C (2014) Integration host factor is required for the induction of acid resistance in Escherichia coli. Curr Microbiol 69:218–224
Biesmans-Oldehinkel E, Doeven MK, Poolman B (2006) ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett 580:1023–1035
Blethen SL, Boeker ZA, Snell EE (1968) Arginine decarboxylase from Escherichia coli. I. Purification and specificity for substrates and coenzyme. J Biol Chem 243:1671–1677
Blum PH, Jovanovich SB, McCann MP, Schultz JE, Lesley SA, Burgess RR, Matin A (1990) Cloning and in vivo and in vitro regulation of cyclic AMP-dependent carbon starvation genes from Escherichia coli. J Bacteriol 172:3813–3820
Bouvier J, Patte JC, Stragier P (1984) Multiple regulatory signals in the control region of the Escherichia coli carAB operon. Proc Natl Acad Sci USA 81:4139–4143
Bouvier J, Stragier P, Morales V, Rémy E, Gutierrez C (2008) Lysine represses transcription of Escherichia coli dapB gene by preventing its activation by the ArgP activator. J Bacteriol 190:5224–5229
Boyen A, Charlier D, Crabeel M, Cunin R, Palchaudhuri S, Glansdorff N (1978) Studies on the control region of the bipolar argECBH operon of Escherichia coli. Mol Gen Genet 161:185–196
Boyen A, Charlier D, Charlier J, Sakanyan V, Mett I, Glansdorff N (1992) Acetylornithine deacetylase, succinyldiaminopimelate desuccinylase and carboxypeptidase G2 are evolutionary related. Gene 116:1–6
Brennan RG (1993) The winged-helix DNA-binding motif: another helix-turn-helix takeoff. Cell 74:773–776
Bretcher AP, Baumberg S (1976) Divergent transcription of the argECBH cluster of Escherichia coli K-12. Mutations which alter the control of enzyme synthesis. J Mol Biol 102:205–220
Brinkman AB, Ettema TJG, de Vos WM, van der Oost J (2003) The Lrp family of transcription regulators. Mol Microbiol 48:287–294
Bröer S, Krämer R (1991a) Lysine excretion by Corynebacterium glutamicum. 1. Identification of a specific secretion carrier system. Eur J Biochem 202:131–135
Bröer S, Krämer R (1991b) Lysine excretion by Corynebacterium glutamicum. 2. Energetics and mechanism of the transport system. Eur J Biochem 202:137–143
Burke M, Merican AF, Sherratt DJ (1994) Mutant Escherichia coli arginine repressor proteins that fail to bind l-arginine, yet retain the ability to bind their normal DNA-binding sites. Mol Microbiol 13:609–618
Burkovski A, Krämer R (2002) Bacterial amino acid transport proteins: occurrence, function, and significance for biotechnological applications. Appl Microbiol Biotechnol 58:265–274
Caldara M, Charlier D, Cunin R (2006) The arginine regulon of Escherichia coli: whole-system transcriptome analysis discovers new genes and provides an integrated view of arginine regulation. Microbiology 152:3343–3354
Caldara M, Nguyen Le Minh P, Bostoen S, Massant J, Charlier D (2007) ArgR-dependent repression of arginine and histidine transport genes in Escherichia coli K-12. J Mol Biol 373:251–267
Caldara M, Dupont G, Leroy F, Goldbeter A, De Vuyst L, Cunin R (2008) Arginine biosynthesis in Escherichia coli: experimental perturbation and mathematical modeling. J Biol Chem 283:6347–6358
Calvo JM, Matthews RG (1994) The Leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev 58:466–490
Canellakis ES, Paterakis AA, Huang SC, Panagiotidis CA, Kyriakidis DA (1993) Identification, cloning, and nucleotide sequencing of the ornithine decarboxylase antizyme gene of Escherichia coli. Proc Natl Acad Sci USA 90:7129–7133
Castanie-Cornet MP, Foster JW (2001) Escherichia coli acid resistance: cAMP receptor protein and a 20-bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709–715
Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW (1999) Control of acid resistance in Escherichia coli. J Bacteriol 181:3525–3535
Castanie-Cornet MP, Cam K, Jacq A (2006) RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli. J Bacteriol 188:4264–4270
Celis TF (1981) Chain-terminating mutants affecting a periplasmic binding protein involved in the active transport of arginine and ornithine in Escherichia coli. J Biol Chem 256:773–779
Celis TF (1982) Mapping of two loci affecting the synthesis and structure of a periplasmic binding protein involved in arginine and ornithine transport in Escherichia coli K-12. J Bacteriol 151:1314–1319
Celis TF, Rosenfeld HJ, Maas WK (1973) Mutant of Escherichia coli defective in transport of basic amino acids. J Bacteriol 116:619–626
Cervera J, Bendala E, Britton HG, Bueso J, Nassif Z, Lusty CJ, Rubio V (1996) Photoaffinity labeling with UMP of lysine 992 of carbamyl phosphate synthetase from Escherichia coli allows identification of the binding sites for the pyrimidine inhibitor. Biochemistry 35:7247–7255
Charlier D, Glansdorff N (2004) Biosynthesis of arginine and polyamines. EcoSal Plus. https://doi.org/10.1128/ecosalplus.3.6.1.10
Charlier D, Crabeel M, Palchaudhuri S, Cunin R, Boyen A, Glansdorff N (1978) Heteroduplex analysis of regulatory mutations and of insertions (IS1, IS2, IS5) in the bipolar argECBH operon of Escherichia coli. Mol Gen Genet 161:175–184
Charlier D, Crabeel M, Cunin R, Glansdorff N (1979) Tandem and inverted repeats of arginine genes in Escherichia coli K12. Mol Gen Genet 174:75–88
Charlier D, Piette J, Glansdorff N (1982) IS3 can function as a mobile promoter in E. coli. Nucleic Acids Res 10:5935–5948
Charlier D, Severne Y, Zafarullah M, Glansdorff N (1983) Turn-on of inactive genes by promoter recruitment in Escherichia coli: inverted repeats resulting in artificial divergent operons. Genetics 105:469–488
Charlier D, Weyens G, Roovers M, Piette J, Bocquet C, Piérard A, Glansdorff N (1988) Molecular interactions in the control region of the carAB operon encoding Escherichia coli carbamoylphosphate synthetase. J Mol Biol 204:867–877
Charlier D, Roovers M, Van Vliet F, Boyen A, Cunin R, Nakamura YG, Glansdorff N, Piérard A (1992) Arginine regulon of Escherichia coli K-12. A study of repressor-operator interactions and of in vitro binding versus in vivo repression. J Mol Biol 226:367–386
Charlier D, Hassanzadeh G, Kholti A, Gigot D, Piérard A, Glansdorff N (1995) carP, involved in pyrimidine regulation of the Escherichia coli carbamoylphosphate synthetase operon encodes a sequence-specific DNA-binding protein identical to XerB and PepA, also required for resolution of ColE1 multimers. J Mol Biol 250:392–406
Charlier D, Nguyen Le Minh P, Roovers M (2018) Regulation of carbamoylphosphate synthesis in Escherichia coli: an amazing metabolite at the crossroad of arginine and pyrimidine biosynthesis. Amino Acids 50:1647–1661
Cherney LT, Cherney MM, Garen CR, Lu GJ, James MNG (2008) Crystal structure of the arginine repressor protein in complex with the DNA operator from Mycobacterium tuberculosis. J Mol Biol 384:1330–1340
Cherney LT, Cherney MM, Garen CR, James MNG (2009) The structure of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator and co-repressor, l-arginine. J Mol Biol 388:85–97
Cho B, Barrett CL, Knight EM, Park YS, Palsson BO (2008) Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc Natl Acad Sci USA 105:19462–19467
Cho BK, Federowicz S, Park YS, Zengler K, Palsson BO (2011) Deciphering the transcriptional regulatory logic of amino acid metabolism. Nat Chem Biol 8:65–71
Cho S, Cho YB, Kang TJ, Kim SC, Palsson BO, Cho BK (2015) The architecture of ArgR-DNA complexes at the genome-scale in Escherichia coli. Nucleic Acids Res 43:3079–3088
Commichau M, Stühlke J (2008) Trigger enzymes: bifunctional enzymes active in metabolism and in controlling gene expression. Mol Microbiol 67:692–702
Crabeel M, Abadjieva A, Hilven P, Desimpelaere J, Soetens O (1997) Characterization of the Saccharomyces cerevisiae ARG7 gene encoding ornithine acetyltransferase, an enzyme with acetylglutamate synthase activity. Eur J Biochem 250:232–241
Cunin R, Glansdorff N (1971) messenger RNA from arginine and phosphoenolpyruvate carboxylase genes in argR + and argR − strains of Escherichia coli K12. FEBS Lett 18:135–137
Cunin R, Elseviers D, Sand G, Freundlich G, Glansdorff N (1969) On the functional organization of the argECBH cluster of genes in Escherichia coli K12. Mol Gen Genet 106:32–47
Cunin R, Boyen A, Pouwels P, Glansdorff N, Crabeel M (1975) Parameters of gene expression in the bipolar argECBH operon of E. coli K12. The question of translational control. Mol Gen Genet 140:51–60
Cunin R, Kelker N, Boyen A, Lang-Yang H, Zubay G, Glansdorff N, Maas WK (1976) Involvement of arginine in in vitro transcription of arginine genen C, B and H in Escherichia coli K-12. Biochem Biophys Res Commun 69:377–382
Cunin R, Eckhardt T, Piette J, Boyen A, Piérard A, Glansdorff N (1983) Molecular basis for modulated regulation of gene expression in the arginine regulon of Escherichia coli K-12. Nucleic Acids Res 11:5007–5019
Cunin R, Glansdorff N, Piérard A, Stalon V (1986) Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev 50:314–352
Czerwinsky RM, Mareya SM, Raushel FM (1995) Regulatory changes in the control of carbamoyl phosphate synthetase induced by truncation and mutagenesis of the allosteric binding domain. Biochemistry 34:13920–13927
Danchin A (2009) Cells need security valves. BioEssays 31:769–773
Davidson AL, Chen J (2004) ATP-binding cassette transporters in bacteria. Annu Rev Biochem 73:241–268
Davidson AL, Dassa E, Orelle C, Chen J (2008) Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72:317–364
De Biase D, Tramonti A, Bossa F, Visca P (1999) The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol Microbiol 32:1198–1211
Delannay S, Charlier D, Tricot C, Villeret V, Piérard A, Stalon V (1999) Serine 948 and threonine 1042 are crucial residues for allosteric regulation of Escherichia coli carbamoylphosphate synthetase and illustrate coupling effects of activation and inhibition pathways. J Mol Biol 286:1217–1228
Devroede N (2006) Mechanisms of purine- and pyrimidine-dependent repression of the Escherichia coli carAB operon, encoding carbamoylphosphate synthase. Dissertation. Vrije Universiteit Brussel
Dohi M, Kikuchi A, Gorini L (1978) Some regulation profiles of ornithine transcarbamylase synthesis in vitro. J Biochem 84:1401–1409
Doroshenko V, Airich L, Vitushkina M, Kolokolova A, Livshits V, Mashko S (2007) YddG from Escherichia coli promotes export of aromatic amino acids. FEMS Microbiol Lett 275:312–318
Dyda F, Klein DC, Hickman AB (2000) GCN5-related N-acetyltransferases: a structural overview. Annu Rev Biophys Biomol Struct 29:81–103
Eckhardt T (1980) Isolation of plasmids carrying the arginine repressor gene argR of Escherichia coli K12. Mol Gen Genet 178:447–452
Eckhardt T, Leisinger T (1975) Isolation and characterization of mutants with a feedback resistant N-acetylglutamate synthase in Escherichia coli K 12. Mol Gen Genet 138:225–232
Eggeling L, Sahm H (2003) New ubiquitous translocators: amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch Microbiol 180:155–160
Elseviers D, Cunin R, Glansdorff N, Baumberg S, Ashcroft E (1972) Control regions within the argECBH cluster of Escherichia coli K12. Mol Gen Genet 117:349–366
Fang Y, Kolmakova-Partensky L, Miller C (2007) A bacterial arginine-agmatine exchange transporter involved in extreme acid resistance. J Biol Chem 282:176–182
Fang Y, Jayaram H, Shane T, Kolmakova-Partensky L, Wu F, Williams C, Xiong Y, Miller C (2009) Structure of a prokaryotic virtual proton pump at 3.2 Å resolution. Nature 460:1040–1043
Fichman Y, Gerdes SY, Kovács H, Szabados L, Zilberstein A, Csonka LN (2015) Evolution of proline biosynthesis: enzymology, bioinformatics, genetics, and transcriptional regulation. Biol Rev 90:1065–1099
Fondi M, Brilli M, Emiliani G, Paffetti D, Fani R (2007) The primordial metabolism: an ancestral interconnection between leucine, arginine and lysine biosynthesis. BMC Evol Biol 7(suppl 2):S3. https://doi.org/10.1186/1471-2148-7-s2-s3
Ford RC, Beis K (2019) Learning the ABCs one at a time: structure and mechanism of ABC transporters. Biochem Soc Trans 47:23–36. https://doi.org/10.1042/bst20180147
Foster JW (2004) Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol 2:898–907
Fraley CD, Kim JH, McCann MP, Matin A (1998) The Escherichia coli starvation gene cstC is involved in amino acid catabolism. J Bacteriol 180:4287–4290
Franke I, Resch A, Daßler T, Maier T, Böck A (2003) YfiK from Escherichia coli promotes export from O-acetylserine and cysteine. J Bacteriol 185:1161–1166
Fröhlich KS, Papenfort K, Berger AA, Vogel J (2012) A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res 40:3623–3640
Fröhlich KS, Haneke K, Papenfort K, Vogel J (2016) The target spectrum of SdsR small RNA in Salmonella. Nucleic Acids Res 44:10406–10422
Gao X, Lu F, Zhou L, Dang S, Sun L, Li X, Wang J, Shi Y (2009) Structure and mechanism of an amino acid antiporter. Science 324:1565–1568
Gao X, Zhou L, Jiao X, Lu F, Yan C, Zeng X, Wang J, Shi Y (2010) Mechanism of substrate recognition and transport by an amino acid antiporter. Nature 463:828–832
Garnett JA, Marincs F, Baumberg S, Stockley PG, Phillips SEV (2008) Structure and function of the arginine repressor-operator complex from Bacillus subtilis. J Mol Biol 379:284–298
Gerosa L, Kochanowski K, Heinemann K, Sauer U (2013) Dissecting specific and global transcriptional regulation of bacterial gene expression. Mol Syst Biol 9:658. https://doi.org/10.1038/msb.2013.14
Gigot D, Crabeel M, Feller A, Charlier D, Lissens W, Glansdorff N, Piérard A (1980) Patterns of polarity in the Escherichia coli carAB gene cluster. J Bacteriol 143:914–920
Ginesy M, Belotserkovsky J, Emman J, Isaksson L, Rova U (2015) Metabolic engineering of Escherichia coli for enhanced arginine biosynthesis. Microb Cell Fact 14:29. https://doi.org/10.1186/s12934-015-0211-y
Glansdorff N (1965) Topography of cotransducible arginine mutations. Genetics 51:167–179
Glansdorff N, Sand G, Verhoef C (1967) The dual genetic control of ornithine transcarbamylase synthesis in Escherichia coli K12. Mutation Res 4:743–751
Glykos NM, Holzenburg A, Phillips SEV (1998) Low-resolution structural characterization of the arginine repressor/activator from Bacillus subtilis: a combined X-ray crystallographic and electron microscopical approach. Acta Cryst D54:215–225
Gong S, Richard H, Foster JW (2003) YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J Bacteriol 185:4402–4409
Gong S, Ma Z, Foster JW (2004) The Era-like GTPase TrmE conditionally activates gadE and glutamate-dependent acid resistance in Escherichia coli. Mol Microbiol 54:948–961
Gorini L, Gundersen W (1961) Induction by arginine of enzymes of arginine biosynthesis in Escherichia coli B. Proc Natl Acad Sci USA 47:961
Gorini L, Gundersen W, Burger M (1961) Genetics of regulation of enzyme synthesis in the arginine biosynthetic pathway of Escherichia coli. Cold Spring Harbor Symp Quant Biol 26:173–182
Goss TJ (2008) The ArgP protein stimulates Klebsiella pneumoniae gdhA promoter in a lysine-sensitive manner. J Bacteriol 190:4351–4359
Grandori R, Lavoie TA, Pflumm M, Tian G, Niersbach H, Maas WK, Fairman R, Carey J (1995) The DNA-binding domain of hexameric arginine repressor. J Mol Biol 254:150–162
Haas D, Kurer V, Leisinger T (1972) N-acetylglutamate synthetase of Pseudomonas aeruginosa. An assay in vitro and feedback inhibition by arginine. Eur J Biochem 31:290–295
Han JS, Kwon HS, Yim JB, Hwang DS (1998) Effect of IciA protein on the expression of the nrd gene encoding ribonucleoside diphosphate reductase in Escherichia coli. Mol Gen Genet 259:610–614
Hayashi SI, Murakami Y, Matsufuji S (1996) Ornithine decarboxylase antizyme: a novel type of regulatory protein. Trends Biochem Sci 21:27–30
He A, Penix SR, Basting RP, Griffith JM, Creaper KE, Camperchioli D, Clark MW, Gonzales AS, Chávez-Eraso J, George NS, Bhagwat AA, Slonczewski JL (2017) Acid evolution of Escherichia coli K-12 eliminates amino acid decarboxylases and reregulates catabolism. Appl Environ Microbiol 83:442. https://doi.org/10.1128/aem.00442-17
Heuveling J, Frochaux V, Ziomkowska J, Wawrzinek R, Wessig P, Herrmann A (2014) Schneider E (2014) Conformational changes of the bacterial type I ATP-binding cassette importer HisQMP2 at distinct steps of the catalytic cycle. Biochim Biophys Acta 1838(1 Pt B):106–116. https://doi.org/10.1016/j.bbamem.2013.08.024
Higgins CF, Ames GFL (1981) Two periplasmic transport proteins which interact with a common membrane protein show extensive homology: complete nucleotide sequence sequences. Proc Natl Acad Sci USA 78:6038–6042
Holden HM, Thoden JB, Raushel FM (1999) Carbamoyl phosphate synthetase: an amazing biochemical odyssey from substrate to product. Cell Mol Life Sci 56:507–522
Hommais F, Krin E, Laurent-Winter C, Soutouria O, Malpertuy A, Le Caer JP, Danchin A, Bertin P (2001) Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol Microbiol 40:20–36
Hosie A, Poole P (2001) Bacterial ABC transporters of amino acids. Res Microbiol 152:259–270
Houghton JE, Bencini DA, O’Donovan GA, Wild JR (1984) Protein differentiation: a comparison of aspartate transcarbamoylase and ornithine transcarbamoylase from Escherichia coli K12. Proc Natl Acad Sci USA 81:4864–4868
Hu M, Deonier C (1981) Mapping IS elements flanking the argF region of the Escherichia coli K-12 chromosome. Mol Gen Genet 181:222–229
Hung SP, Baldi P, Hatfield GW (2002) Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J Biol Chem 277:40309–40323
Hwang DS, Kornberg A (1990) A novel protein binds a key origin sequence to block replication of an E. coli minichromosome. Cell 63:325–331
Hwang DS, Kornberg A (1992) Opposed actions of regulatory proteins, DnaA and IciA, in opening the replication origin of Escherichia coli. J Biol Chem 267:23087–23091
Hwang DS, Tony B, Kornberg A (1992) IciA protein, a specific inhibitor of initiation of Escherichia coli chromosomal replication. J Biol Chem 267:2209–2213
Igarashi K, Kashiwagi K (2018) Effects of polyamines on protein synthesis and growth of Escherichia coli. J Biol Chem 293:18702–18709
Ikeda M (2003) Amino acid production processes. Adv Biochem Eng Biotechnol 79:1–35
Ikeda M, Mitsuhashi S, Tanaka K, Hayashi M (2009) Reengineering of Corynebacterium glutamicum l-arginine and l-citrulline producers. Appl Environ Microbiol 75:1635–1641
Ilgü H, Jeckelmann JM, Gapsys V, Ucurum Z, de Groot BL, Fotiadis D (2016) Insights into the molecular basis for substrate binding and specificity of the wild-type l-arginine/agmatine antiporter AdiC. Proc Natl Acad Sci USA 113:10358–10363
Ishihama A (2010) Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchical networks. FEMS Microbiol Rev 34:628–645
Ishihama A, Kori A, Koshio E, Yamada K, Maeda H, Shimada T, Makinoshima H, Iwata A, Fujita N (2014) Intracellular concentrations of 65 species of transcription factors with known regulatory functions in Escherichia coli. J Bacteriol 196:2718–2727
Itikawa H, Baumberg S, Vogel H (1968) Enzymatic basis for a genetic suppression: accumulation and deacylation of N-acetylglutamine-γ-semialdehyde in enteric bacterial mutants. Biochim Biophys Acta 159:547–550
Itoh Y (1997) Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa. J Bacteriol 179:7280–7290
Iyer R, Williams C, Miller C (2003) Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J Bacteriol 185:6556–6561
Jack DL, Paulsen IT, Saier MH (2000) The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146:1797–1814
Jacoby GA (1971) Mapping the gene determining ornithine transcarbamylase and its operator in Escherichia coli B. J Bacteriol 108:645–651
Jacoby GA (1972) Control of the argECBH cluster in Escherichia coli. Mol Gen Genet 117:337–348
Jann A, Matsumoto H, Haas D (1988) The fourth arginine catabolic pathway of Pseudomonas aeruginosa. J Gen Microbiol 134:1043–1053
Javid-Majd F, Blanchard JS (2000) Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase. Biochemistry 39:1285–1293
Jin L, Xue WF, Fukayama JW, Yetter J, Pickering M, Carey J (2005) Assymetric allosteric activation of the symmetric ArgR hexamer. J Mol Biol 346:43–56
Jones ME, Spector L, Lipmann F (1955) Carbamyl phosphate, the carbamyl donor in enzymatic citrulline synthesis. J Am Chem Soc 77:819–820
Jones CM, Hernández Lozada NJ, Pfleger BF (2015) Efflux systems in bacteria and their metabolic engineering applications. Appl Microbiol Biotechnol 99:9381–9393
Jung H, Pirch T, Hilger D (2006) Secondary transport of amino acids in prokaryotes. J Membr Biol 213:119–133
Kanjee U, Houry WA (2013) Mechanisms of acid resistance in Escherichia coli. Annu Rev Microbiol 67:65–81
Kashiwagi K, Suzuki T, Suzuki F, Furuchi T, Kobayashi H, Igarashi K (1991) Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J Biol Chem 266:20922–20927
Kashiwagi K, Miyamoto S, Suzuki F, Kobayashi H, Igarashi K (1992) Excretion of putrescine by the putrescine-ornithine antiporter encoded by the potE gene of Escherichia coli. Proc Natl Acad Sci USA 89:4529–4533
Kashiwagi K, Shibuya S, Tomitori H, Kuraishi A, Igarashi K (1997) Excretion and uptake of putrescine by the PotE protein of Escherichia coli. J Biol Chem 272:6318–6323
Kashiwagi K, Kuraishi A, Tomitori H, Igarashi A, Nishimura K, Shirahata A, Igarashi K (2000) Identification of the putrescine recognition site on polyamine transport protein PotE. J Biol Chem 275:36007–36012
Kawashima T, Aramaki H, Oyamada T, Makino K, Yamada M, Okamura H, Yokoyama K, Ishijima SA, Suzuki M (2008) Transcription regulation by feast/famine regulatory proteins, FFRPs, in Archaea and Eubacteria. Biol Pharm Bull 31:173–186
Kelker N, Maas WK (1974) Selection of genetically repressible (argR +) strains of E. coli K-12 from genetically derepressed (argR −) mutants using acetylnorvaline. Mol Gen Genet 132:131–135
Kelker N, Maas WK, Yang HL, Zubay G (1976) In vitro synthesis and repression of argninosuccinase in Escherichia coli K-12: partial purification of the arginine repressor. Mol Gen Genet 144:17–20
Kelln RA, O’Donovan GA (1976) Isolation and partial characterization of an argR mutant of Salmonella typhimurium. J Bacteriol 128:528–535
Kerppola RE, Shyamala VK, Klebba P, Ames GFL (1991) The membrane-bound proteins of periplasmic permeases form a complex. Identification of the histidine permease HisQMP complex. J Biol Chem 266:9857–9865
Kholti A, Charlier D, Gigot D, Huysveld N, Roovers M, Glansdorff N (1998) pyrH-encoded UMP-kinase directly participates in pyrimidine-specific modulation of promoter activity in Escherichia coli. J Mol Biol 280:571–582
Kilstrup M, Lu CD, Abdelal A, Neuhard J (1988) Nucleotide sequence of the carA gene and regulation of the carAB operon in Salmonella typhimurium. Eur J Biochem 176:421–429
Kiupakis AK, Reitzer LJ (2002) ArgR-independent induction and ArgR-dependent superinduction of the astCADBE operon in Escherichia coli. J Bacteriol 184:2940–2950
Kowalczyk I, Ratera M, Paladino A, Bartoccioni P, Errasti-Murugarren F, Valencia E, Portella G, Bial S, Zorzano A, Fita I, Orozco M, Carpena X, Vázquez-Ibar JL, Palacín M (2011) Molecular basis of substrate-induced permeation by an amino acid antiporter. Proc Natl Acad Sci USA 108:3935–3940
Krämer R (1994) Systems and mechanisms of amino acid uptake and excretion in prokaryotes. Arch Microbiol 162:1–13
Krin E, Laurent-Winter C, Bertin PN, Danchin A, Kolb A (2003) Transcription regulation coupling of the divergent argG and metY promoters in Escherichia coli K-12. J Bacteriol 185:3139–3146
Krzyzek RA, Rogers P (1976) Effect of arginine on the stability and size of argECBH messenger ribonucleic acid in Escherichia coli. J Bacteriol 126:365–376
Kueh R, Rahman NA, Merican AF (2003) Computational docking of l-arginine and its structural analogues to C-terminal domain of Escherichia coli arginine repressor (ArgRc). J Mol Model 9:88–98
Kuo T, Stocker BAD (1969) Suppression of proline requirement of proA and proAB deletion mutants in Salmonella typhimurium by mutation to arginine requirement. J Bacteriol 98:593–598
Kustu SG, Ames GFL (1973) The HisP protein, a known histidine transport component in Salmonella typhimurium, is also an arginine transport component. J Bacteriol 116:107–113
Kutukova EA, Livshits VA, Altman IA, Ptitsyn LR, Zyiatdinov MH, Tokmakova IL, Zakataeva NP (2005) The yeaS (leuE) gene of Escherichia coli encodes an exporter of leucine, and the Lrp protein regulates its expression. FEBS Lett 579:4629–4634
Labedan B, Boyen A, Baetens M, Charlier D, Chen P, Cunin R, Durbecq V, Glansdorff N, Herve G, Legrain C, Liand Z, Purcarea C, Roovers M, Sanchez R, Toong TL, Van de Casteele M, Van Vliet F, Xu Y, Zhang YF (1999) The evolutionary history of carbamoyltransferases: a complex set of paralogous genes was already present in the last universal common ancestor. J Mol Evol 49:461–473
Lacour S, Landini P (2004) σS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of σS-dependent genes and identification of their promoter sequences. J Bacteriol 186:7186–7195
Laishram RS, Gowrishankar J (2007) Environmental regulation operating at the promoter clearance step of bacterial transcription. Genes Dev 21:1258–1272
Lal PB, Schneider BL, Vu K, Reitzer L (2014) The redundant aminotransferases in lysine and arginine synthesis and the extent of aminotransferase redundancy in Escherichia coli. Mol Microbiol 94:843–856
Law CJ, Maloney PC, Wang DN (2008) Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol 62:289–305
Ledwige R, Blanchard JS (1999) The dual biosynthetic capability of N-acetylornithine aminotransferase in arginine and lysine biosynthesis. Biochemistry 38:3019–3024
Lee Y, Lee H, Yim J, Hwang DS (1997) The binding of two dimers of IciA protein to the dnaA promoter 1P element enhances the binding of RNA polymerase to the dnaA promoter 1P. Nucleic Acids Res 25:3486–3489
Legrain C, Halleux P, Stalon V, Glansdorff N (1972) The dual genetic control of ornithine carbamoyltransferase in Escherichia coli: a case of bacterial hybrid enzymes. Eur J Biochem 27:93–102
Legrain C, Stalon V, Glansdorff N (1976) Escherichia coli ornithine carbamoyltransferase isoenzymes: evolutionary significance and the isolation of λargF and λargI transducing phages. J Bacteriol 128:35–38
Leisinger T, Haas D (1975) N-acetylglutamate synthetase of Escherichia coli: regulation of synthesis and activity by arginine. J Biol Chem 250:1690–1693
Lejeune D, Delsaux N, Charloteaux B, Thomas A, Brasseur T (2005) Protein-nucleic acid recognition: statistical analysis of atomic interactions and influence of DNA structure. Proteins 61:258–271
Lim D, Oppenheim JD, Eckhardt T, Maas WK (1987) Nucleotide of the argR gene of Escherichia coli K12 and isolation of its product the arginine repressor. Proc Natl Acad Sci USA 84:6697–6701
Lim D, Oppenheim JD, Eckhardt T, Maas WK (1988) The unitary hypothesis for the repression mechanism of arginine biosynthesis in E. coli B and E. coli K12—revisited after 18 years. In Symposium on Gene Expression and Regulation: the Legacy of Luigi Gorini. Elsevier Science Publishers, Amsterdam
Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW (1995) Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol 177:4097–4104
Lipscomb WN, Kantrowitz ER (2012) Structure and mechanisms of Escherichia coli aspartate transcarbamoylase. Acc Chem Res 45:444–453
Lissens W, Cunin R, Kelker N, Glansdorff N, Piérard A (1980) In vitro synthesis of Escherichia coli carbamoylphosphate synthase: evidence for a participation of the arginine repressor in cumulative repression. J Bacteriol 141:58–66
Livshits VA, Zakaraeva NP, Aleshin VV, Vitushkina MV (2003) Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res Microbiol 154:123–135
Lu CD (2006) Pathways and regulation of bacterial arginine metabolism and perspectives for obtaining arginine overproducing strains. App Microbiol Biotechnol 70:261–272
Lu CD, Abdelal AT (1999) Role of ArgR in activation of the ast operon, encoding enzymes of the arginine succinyltransferase pathway in Salmonella typhimurium. J Bacteriol 181:1934–1938
Lu CD, Houghton JE, Abdelal AT (1992) Characterization of the arginine repressor from Salmonella typhimurium and its interaction with the carAB operator. J Mol Biol 225:11–24
Lustig B, Jernigan RL (1995) Consistencies of individual DNA base-amino acid interactions in structures and sequences. Nucleic Acids Res 23:4707–4711
Ma Z, Richard H, Tucker D, Conway T, Foster JW (2002) Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YihW). J Bacteriol 184:7001–7012
Ma Z, Gong S, Richard H, Tucker D, Conway T, Foster JW (2003a) GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol Microbiol 49:1309–1320
Ma Z, Richard H, Foster JW (2003b) pH-Dependent modulation of cyclic AMP levels and GadW-dependent repression of RpoS affect synthesis of GadX regulator and Escherichia coli acid resistance. J Bacteriol 185:6852–6859
Ma Z, Masuda N, Foster JW (2004) Characterization of EvgAS-YdeO-GadE branched regulatory circuit governing glutamate-dependent acid resistance in Escherichia coli. J Bacteriol 186:7378–7389
Maas WK (1957) Regulation of arginine biosynthesis in Escherichia coli. Biol Bull 111:319
Maas WK (1961) Studies on repression of arginine biosynthesis in Escherichia coli. Cold Spring Harbor Symp Quant Biol 26:183–191
Maas WK (1991) The regulation of arginine biosynthesis: its contribution to understanding the control of gene expression. Genetics 128:489–494
Maas WK (2007) The potential for the formation of the arginine biosynthetic enzymes and its masking during evolution. BioEssays 29:484–488
Maas WK, Clark AJ (1964) Studies on the mechanism of repression of arginine biosynthesis in Escherichia coli. II. Dominance of repressibility in diploids. J Mol Biol 8:365–370
Maddocks SE, Oyston PCF (2008) Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623
Marbaniang CN, Gowrishankar J (2011) Role of ArgP (IciA) in lysine-mediated repression in Escherichia coli. J Bacteriol 193:5985–5996
Marbaniang CN, Gowrishankar J (2012) Transcriptional cross-regulation between Gram-negative and Gram-positive bacteria, demonstrated using ArgP-argO of Escherichia coli and LysG-lysE of Corynebacterium glutamicum. J Bacteriol 194:5657–5666
Marc F, Weigel P, Legrain C, Almeras Y, Santrot M, Glansdorff N, Sakanyan V (2000) Characterization and kinetic mechanism of mono- and bifunctional ornithine acetyltransferases from thermophilic microorganisms. Eur J Biochem 267:5217–5226
Marc F, Weigel P, Legrain C, Glansdorff N, Sakanyan V (2001) An invariant threonine is involved in self-catalyzed cleavage of the precursor protein for ornithine acetyltransferase. J Biol Chem 276:25404–25410
Marvil DK, Leisinger T (1977) N-acetylglutamate synthase of Escherichia coli. Purification, characterization, and molecular properties. J Biol Chem 252:3295–3303
Masuda N, Church GM (2002) Escherichia coli gene expression responsive to levels of the response regulator EvgA. J Bacteriol 184:6225–6234
Masuda N, Church GM (2003) Regulatory network of acid resistance genes in Escherichia coli. Mol Microbiol 48:699–712
Mergeay M, Gigot D, Beckman J, Glansdorff N, Piérard A (1974) Physiology and genetics of carbamoylphosphate synthesis in Escherichia coli K-12. Mol Gen Genet 133:299–316
Merlo LMF, Sadowsky MJ, Ferguson JA, Dean AM (2006) The argR B of Escherichia coli is rare in isolates obtained from natural sources. Gene 376:240–247
Michael AJ (2018) Polyamine function in archaea and bacteria. J Biol Chem 293:18693–18701
Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M (2015) Remaining mysteries of molecular biology: the role of polyamines in the cell. J Mol Biol 427:3389–3406
Miltcheva Karaivanova I, Weigel P, Takahashi M, Fort C, Versavaud A, Van Duyne G, Charlier D, Hallet JN, Glansdorff N, Sakanyan V (1999) Mutational analysis of the thermostable arginine repressor from Bacillus stearothermophilus: dissecting residues involved in DNA binding properties. J Mol Biol 291:843–855
Minh PN, Devroede N, Massant J, Maes D, Charlier D (2009) Insights into the architecture and stoichiometry of Escherichia coli PepA·DNA complexes involved in transcriptional control and site-specific DNA recombination by atomic force microscopy. Nucleic Acids Res 37:1463–1476
Miyazaki J, Kobashi N, Nishiyama M, Yamane H (2001) Functional and evolutionary relationship between arginine biosynthesis and prokaryotic lysine biosynthesis through α-aminoadipate. J Bacteriol 183:5067–5073
Monsieurs P, De Keersmaecker S, Navarre WW, Bader MW, De Smet F, McClelland M, Fang FC, De Moor B, Vanderleyden J, Marchal K (2005) Comparison of the PhoPQ regulon in Escherichia coli and Salmonella typhimurium. J Mol Evol 60:462–474
Morizono H, Cabrera-Luque J, Shi D, Gallegos R, Yamaguchi S, Yu X, Allewell N, Malamy M, Tuchman M (2006) Acetylornithine transcarbamylase: a novel enzyme in arginine biosynthesis. J Bacteriol 188:2974–2982
Morris DR, Boeker FA (1983) Biosynthetic and biodegradative ornithine and arginine decarboxylases from Escherichia coli. Methods Enzymol 94:125–134
Morris DR, Pardee AB (1965) A biosynthetic ornithine decarboxylase in Escherichia coli. Biochem Biophys Res Commun 20:697–702
Nandineni MR, Gowrishankar J (2004) Evidence for an arginine exporter encoded by yggA (argO) that is regulated by the LysR-type transcriptional regulator ArgP in Escherichia coli. J Bacteriol 186:3539–3546
Nandineni MR, Laishram RS, Gowrishankar J (2004) Osmosensitivity associated with insertions in argP (iciA) or glnE in glutamate synthase-deficient mutants of Escherichia coli. J Bacteriol 186:6391–6399
Newman E, Lin R (1995) Leucine responsive regulatory protein: a global regulator for gene expression in E. coli. Ann Rev Microbiol 49:747–775
Nguyen Le Minh P, Nadal M, Charlier D (2016) The trigger enzyme PepA (aminopeptidase A) of Escherichia coli, a transcriptional repressor that generates positive supercoiling. FEBS Lett 590:1816–1825
Nguyen Le Minh P, Ruiz Velázquez, Vandermeeren S, Abwoyo P, Bervoets I, Charlier D (2018) Differential protein-DNA contacts for activation and repression by ArgP, a LysR-type (LTTR) transcriptional regulator in Escherichia coli. Microbiol Res 206:141–158
Ni J, Sakanyan V, Charlier D, Glansdorff N, Van Duyne GD (1999) Structure of the arginine repressor from Bacillus stearothermophilus. Nat Struct Biol 6:427–432
Niersbach H, Lin R, Van Duyne GD, Maas WK (1998) A superrepressor mutant of the arginine repressor with a correctly predicted alteration of ligand binding specificity. J Mol Biol 279:753–760
Nishida H, Nishiyama M, Kobashi N, Kosuge T, Hoshino T, Yamane H (1999) A prokaryotic gene cluster involved in synthesis of lysine through the amino adipate pathway: a key to the evolution of amino acid biosynthesis. Genome Res 9:1175–1183
Panagiotidis CA, Huang SC, Cannelakis ES (1994) Post-translational and transcriptional regulation of polyamine biosynthesis is Escherichia coli. Int J Biochem 26:991–1001
Panagiotidis CA, Huang SC, Cannelakis ES (1995) Relationship of the expression of the S20 and L34 ribosomal proteins to polyamine biosynthesis in Escherichia coli. Int J Biochem Cell Biol 27:157–168
Panchal CJ, Bagchee SN, Guha A (1974) Divergent orientation of transcription from the argECBH operon of Escherichia coli. J Bacteriol 117:675–680
Pannekoek H, Cunin R, Boyen A, Glansdorff N (1975) In vitro transcription of the bipolar argECBH operon in Escherichia coli K12. FEBS Lett 51:143–145
Pathania A, Sardesai AA (2015) Distinct paths for basic amino acid export in Escherichia coli: YbjE (LysO) mediates export of l-lysine. J Bacteriol 197:2036–2047
Pathania A, Gupta AK, Dubey S, Gopal B, Sardesai AA (2016) The topology of the l-arginine exporter ArgO conforms to an Nin-Cout configuration in Escherichia coli: requirement for the cytoplasmic N-terminal domain, functional helical interaction, and an aspartate pair for ArgO function. J Bacteriol 198:3186–3199
Pauwels K, Abadjieva A, Hilven P, Stanckievicz A, Crabeel M (2003) The N-acetylglutamate synthase/N-acetylglutamate kinase metabolon of Saccharomyces cerevisiae allows co-ordinated feedback regulation of the first two steps in arginine biosynthesis. Eur J Biochem 270:1014–1024
Peeters E, Nguyen Le Minh P, Foulquié-Moreno M, Charlier D (2009) Competitive activation of the Escherichia coli argO gene coding for an arginine exporter by the transcriptional regulators Lrp and ArgP. Mol Microbiol 74:1513–1526
Piérard A (1966) Control of the activity of Escherichia coli carbamoylphosphate synthetase by antagonistic allosteric effectors. Science 154:1572–1573
Piérard A, Wiame JM (1964) Regulation and mutation affecting a glutamine-dependent formation of carbamoylphosphate in Escherichia coli. Biochem Biophys Res Commun 15:76–81
Piérard A, Glansdorff N, Mergeay M, Wiame JM (1965) Control of the biosynthesis of carbamoylphosphate in Escherichia coli. J Mol Biol 14:23–36
Piérard A, Lissens W, Halleux P, Cunin R, Glansdorff N (1980) Role of transcriptional regulation and enzyme inactivation in the synthesis of Escherichia coli carbamoylphosphate synthetase. J Bacteriol 141:382–385
Piette J, Cunin R, Boyen A, Charlier D, Crabeel M, Van Vliet F, Glansdorff N, Squires C, Squires CL (1982a) The regulatory region of the divergent argECBH operon in Escherichia coli K12. Nucleic Acids Res 10:8031–8048
Piette J, Cunin R, Van Vliet F, Charlier D, Crabeel M, Ota Y, Glansdorff N (1982b) Homologous control siyes and DNA transcription starts in the related argF and argI genes of Escherichia coli K12. EMBO J 1:853–857
Piette J, Nyunoya H, Lusty CJ, Cunin R, Weyens G, Crabeel M, Charlier D, Glansdorff N, Piérard A (1984) DNA sequence of the carA gene and control region of carAB: tandem promoters, respectively controlled by arginine and the pyrimidines, regulate the synthesis of carbamoylphosphate synthetase in Escherichia coli K12. Proc Natl Acad Sci USA 81:4134–4138
Pouwels PH, Cunin R, Glansdorff N (1974) Divergent transcription in the argECBH cluster of genes in Escherichia coli K12. J Mol Biol 83:421–424
Rajagopal BS, DePonte J 3rd, Tuchman M, Malamy MH (1998) Use of inducible feedback-resistant N-acetylglutamate synthetase (argA) genes for enhanced arginine biosynthesis by genetically engineered Escherichia coli K-12 strains. Appl Envir Microbiol 64:1805–1811
Ramon-Maiques S, Marina A, Gil-Ortiz F, Fita I, Rubio V (2002) Structure of acetylglutamate kinase, a key enzyme for arginine biosynthesis and a prototype for the amino acid kinase enzyme family, during catalysis. Structure 10:329–342
Ramon-Maiques S, Fernandez-Murga ML, Gil-Ortiz F, Vagin A, Fita I, Rubio V (2006) Structural bases of feed-back control of arginine biosynthesis, revealed by the structures of two hexameric N-acetylglutamate kinases, from Thermotoga maritima and Pseudomonas aeruginosa. J Mol Biol 356:695–713
Raushel FM, Thoden JB, Holden HM (1999) The amidotransferase family of enzymes: molecular machines for the production and delivery of ammonia. Biochemistry 38:7891–7899
Reitzer LJ (2005) Catabolism of amino acids and related compounds. EcoSal Plus. https://doi.org/10.1128/ecosalplus.3.4.7
Reitzer LJ, Schneider BL (2001) Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol Mol Biol Rev 65:422–444
Rhodes D, Schwabe JWR, Chapman L, Fairall L (2010) Towards an understanding of protein-DNA recognition. Philos Trans R Soc Lond B 351:501–509
Richard H, Foster JW (2004) Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol 186:6032–6041
Rogers P, Krzyzek R, Kaden TM, Arfman E (1971) Effect of arginine and canavanine on arginine messenger RNA synthesis. Biochem Biophys Res Commun 44:1220–1226
Rogers P, Kaden TM, Toth J (1975) Repression of arg mRNA synthesis by l-arginine in cell-free extracts of Escherichia coli. Biochem Biophys Res Commun 65:1284–1291
Roovers M, Charlier D, Feller A, Gigot D, Holemans F, Lissens W, Piérard A, Glansdorff N (1988) carP, a novel gene regulating the transcription of the carbamoylphosphate synthetase operon of Escherichia coli. J Mol Biol 204:857–865
Rosen BP (1971) Basic amino acid transport in Escherichia coli. J Biol Chem 246:3653–3662
Rubio V, Cervera J, Lusty CJ, Bendala E, Britton HG (1991) Domain structure of the large subunit of Escherichia coli carbamoyl phosphate synthetase. Location of the binding site for the allosteric inhibitor UMP in the COOH-terminal domain. Biochemistry 30:1068–1075
Ruiz J, Haneburger I, Jung K (2011) Identification of ArgP and Lrp as transcriptional regulators of lysP, the gene encoding the specific lysine permease of Escherichia coli. J Bacteriol 193:2536–2548
Saier HJ (2000) Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology 146:1775–1795
Sakanyan V, Kochikyan A, Mett I, Legrain C, Charlier D, Piérard A, Glansdorff N (1992) A re-examination of the pathway for ornithine biosynthesis in a thermophilic and two mesophilic Bacillus species. J Gen Microbiol 138:125–130
Sakanyan V, Charlier D, Legrain C, Kochikyan A, Mett I, Piérard A, Glansdorff N (1993a) Primary structure, partial purification and regulation of key enzymes of the acetyl cycle of arginine biosynthesis in Bacillus stearothermophilus: dual function of ornithine acetyltransferase. J Gen Microbiol 139:393–402
Sakanyan V, Desmarez M, Legrain C, Charlier D, Mett I, Kochikyan A, Savchenko A, Boyen A, Falmagne P, Piérard A, Glansdorff N (1993b) Gene cloning, sequence analysis, purification, and characterization of a thermostable aminoacylase from Bacillus stearothermophilus. Appl Environ Microbiol 59:3878–3888
Sakanyan V, Petrosyan P, Lecocq M, Boyen A, Legrain C, Demarez M, Hallet JN, Glansdorff N (1996) Genes and enzymes of the acetyl cycle of arginine biosynthesis in Corynebacterium glutamicum: enzyme evolution in the early steps of the arginine pathway. Microbiology 142(Pt 1):99–108
Samalíková M, Carey J, Grandori R (2005) Assembly of the hexameric Escherichia coli arginine repressor investigated by nano-electrospray ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 19:2549–2552
Sander T, Farke N, Diehk C, Kuntz M, Glatter T, Link H (2019) Allosteric feedback inhibition enables robust amino acid biosynthesis in E. coli by enforcing enzyme overabundance. Cell Systems 8:66–75
Sayed AK, Odom C, Foster JW (2007) The Escherichia coli AraC-family regulators GadX and GadW activate gadE, the central activator of glutamate-dependent acid resistance. Microbiology 153:2584–2592
Schneefeld M, Busche T, Geffers R, Kalinowski J, Bange FC (2017) The transcriptional regulator LysG (Rv1985c) of Mycobacterium tuberculosis activates lysE (Rv1986) in a lysine-dependent manner. PLoS One 12(10):e0186505. https://doi.org/10.1371/journal.pone.0186505
Schneider BL, Kiupakis AK, Reitzer LJ (1998) Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J Bacteriol 180:4278–4286
Sens D, Natter W, Garvin RT, James E (1977a) Transcription of the argF and argI genes of the arginine biosynthetic regulon of E. coli. Mol Gen Genet 155:7–18
Sens D, Natter W, James E (1977b) In vitro transcription of the Escherichia coli K-12 argA, argE, and argCBH operons. J Bacteriol 130:642–655
Shi X, Bennett GN (1994) Effects of rpoA and cysB mutations on acid induction of biodegradative arginine decarboxylase in Escherichia coli. J Bacteriol 176:7017–7023
Shi X, Waasdorp BC, Bennett GN (1993) Modulation of acid-induced amino acid decarboxylase gene expression by hns in Escherichia coli. J Bacteriol 175:1182–1186
Shi D, Morizono Y, Xiaolin Y, Roth L, Caldovic L, Allewell NM, Tuchman M (2005) Crystal structure of a N-acetylornithine transcarbamylase from Xanthomonas campestris: a novel enzyme in a new arginine biosynthetic pathway found in several eubacteria. J Biol Chem 280:14366–14369
Shi D, Morizono H, Cabrera-Luque J, Xiaolin Y, Roth L, Malamy MH, Allewell NM, Tuchman M (2006) Structure and catalytic mechanism of a novel N-succinyl-l-ornithine transcarbamylase in arginine biosynthesis of Bacteroides fragilis. J Biol Chem 281:20623–20631
Shi D, Caldovic L, Tuchman M (2018) Sources and fates of carbamyl phosphate: a labile energy-rich molecule with multiple facets. Biology 7:E34. https://doi.org/10.3390/biology7020034
Stalon V, Vander Wauven C, Momin P, Legrain C (1987) Catabolism of arginine, citrulline and ornithine by Pseudomonas and related species. J Gen Microbiol 133:2487–2495
Stim-Herndon KP, Flores TM, Bennett GN (1996) Molecular characterization of adiY, a regulatory gene which affects expression of the biodegradative acid-induced arginine decarboxylase gene (adiA) of Escherichia coli. Microbiology 142:1311–1320
Stirling CJ, Szatmari G, Stewart G, Smith CH, Sherratt DJ (1988) The arginine repressor is essential for plasmid stabilizing site-specific recombination at the ColE1 cer locus. EMBO J 7:4389–4395
Stirling CJ, Colloms SD, Collins JF, Szatmari G, Sherratt DJ (1989) xerB, an Escherichia coli gene required for plasmid ColE1 site-specific recombination, is identical to pepA, encoding aminopeptidase A, a protein with substantial similarity to bovine lens leucine aminopeptidase. EMBO J 8:1623–1627
Strawn R, Melicherick M, Green M, Stocker T, Carey J, Ettrich R (2010) Symmetric allosteric mechanism of hexameric Escherichia coli arginine repressor exploits competition between l-arginine ligands and resident arginine residues. PLoS Comput Biol 6(6):e1000801. https://doi.org/10.1371/journal.pcbi.1000801
Suiter AM, Bänziger O, Dean AM (2003) Fitness consequences of a regulatory polymorphism in a seasonal environment. Proc Natl Acad Sci USA 100:12782–12786
Sunnerhagen M, Nilges M, Otting G, Carey J (1997) Solution structure of the DNA-binding domain and model for the complex of multifunctional hexameric arginine repressor with DNA. Nat Struct Biol 4:819–826
Szwajkajzer D, Dai L, Fukayama JW, Abramczyk B, Fairman R, Carey J (2001) Quantitative analysis of DNA binding by the Escherichia coli arginine repressor. J Mol Biol 312:949–962
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36
ter Beek J, Guskov A, Slotboom DJ (2014) Structural diversity of ABC transporters. J Gen Physiol 143:419–435
Thoden JB, Raushel FM, Wesenberg G, Holden HM (1999a) The binding of inosine monophosphate to Escherichia coli carbamoyl phosphate synthetase. J Biol Chem 274:22502–22507
Thoden JB, Wesenberg G, Raushel FM, Holden HM (1999b) Carbamoyl phosphate synthetase: closure of the B-domain as a result of nucleotide binding. Biochemistry 38:2347–2357
Tian G, Maas WK (1994) Mutational analysis of the arginine repressor of Escherichia coli. Mol Microbiol 13:599–608
Tian G, Lim D, Carey J, Maas WK (1992) Binding of the arginine repressor of Escherichia coli to its operator sites. J Mol Biol 226:387–397
Tian G, Lim D, Oppenheim JD, Maas WK (1994) Explanation for different types of regulation of arginine biosynthesis in Escherichia coli B and Escherichia coli K12 caused by a difference between their arginine repressors. J Mol Biol 235:221–230
Tomitori H, Kashiwagi K, Igarashi K (2012) Structure and function of polyamine-amino acid antiporters CadB and PotE in Escherichia coli. Amino Acids 42:733–740
Torres Montaguth OE (2014) Transcriptional regulation of arginine transport genes in Escherichia coli. Dissertation, Vrije Universiteit Brussel
Tramonti A, Visca P, de Canio M, Falconi M, De Biase D (2002) Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J Bacteriol 184:2603–2613
Tricot C, Stalon V, Legrain C (1991) Isolation and characterization of Pseudomonas putida mutants affected in arginine, ornithine and citrulline catabolism: function of the arginine oxidase and arginine succinyltransferase pathways. J Gen Microbiol 137:2911–2918
Tsai MF, Miller C (2013) Substrate selectivity in arginine-dependent acid resistance in enteric bacteria. Proc Natl Acad Sci USA 110:5893–5897
Tuchman M, Rajagopal BS, McCann MT, Malamy MH (1997) Enhanced production of arginine and urea by genetically engineered Escherichia coli K-12 strains. Appl Environ Microbiol 63:33–38
Udaka S (1966) Pathway-specific pattern of control of arginine biosynthesis in bacteria. J Bacteriol 91:617–621
Udaka S (1970) Isolation of the arginine repressor in Escherichia coli. Nature (London) 228:336–338
Udaka S, Kinoshita S (1958) Studies on l-ornithine fermentation. I. The biosynthesis pathway of l-ornithine in Micrococcus glutamicus. J Gen Appl Microbiol 4:283–288
Van de Casteele M, Desmarez M, Legrain C, Piérard A, Glansdorff N (1990) Pathways of arginine biosynthesis in extreme thermophilic archaeo- and eubacteria. J Gen Microbiol 136:1177–1183
Van Duyne GD, Ghosh G, Maas WK, Sigler PB (1996) Structure of the oligomerization and l-arginine binding domain of the arginine repressor of Escherichia coli. J Mol Biol 256:377–391
Van Vliet F, Cunin R, Jacobs A, Piette J, Gigot D, Lauwereys A, Piérard A, Glansdorff N (1984) Evolutionary divergence of genes for ornithine and aspartate carbamoyltransferases-complete sequence and mode of regulation of the Escherichia coli argF gene: comparison with argI and pyrB. Nucleic Acids Res 12:6277–6289
Vander Wauven C, Stalon V (1985) Occurrence of succinyl derivatives in the catabolism of arginine in Pseudomonas cepacia. J Bacteriol 164:882–886
Velasco AM, Leguina JI, Lazcano A (2002) Molecular evolution of the lysine biosynthetic pathways. J Mol Evol 55:445–459
Vetting MW, de Carvalho LPS, Yu M, Hedge SS, Magnet S, Roderick SL, Blanchard JS (2005) Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys 433:212–226
Vogel HJ (1957) Repression and induction as control mechanisms of enzyme biogenesis: the “adaptive” formation of acetylornithinase. In: McElroy WD, Glass B (eds) The chemical basis of heredity. The John Hopkins Press, Baltimore, pp 269–272
Vogel HJ (1961) Aspects of repression in the regulation of enzyme synthesis: pathway-wide control and enzyme-specific response. Cold Spring Harbor Symp Quant Biol 26:163–171
Vogel HJ (1970) Arginine biosynthetic system in Escherichia coli. Methods Enzymol 17A:260–264
Vrljic M, Sahm H, Eggeling L (1996) A new type of transporter with a new type of cellular function: l-lysine export from Corynebacterium glutamicum. Mol Microbiol 22:815–826
Vrljic MG, Garg J, Bellmann A, Wachi S, Freudl R, Malecki MJ, Sahm H, Kozina VJ, Eggeling L, Saier MH (1999) The LysE superfamily: topology of the lysine exporter LysE of Corynebacterium glutamicum, a paradigm for a novel superfamily of transmembrane solute translocators. J Mol Microbiol Biotecnol 1:327–337
Vyas S, Maas WK (1963) Feedback inhibition of acetylglutamate synthase by arginine in Escherichia coli. Arch Biochem Biophys 100:542–546
Wang H, Charlier D, Glansdorff N (1998) The arginine repressor of Escherichia coli K-12 makes direct contacts to minor and major groove determinants of the operator. J Mol Biol 277:805–824
Wang S, Yan R, Zhang X, Chu Q, Shi Y (2014) Molecular mechanism of pH-dependent substrate transport by an arginine-agmatine antiporter. Proc Natl Acad Sci USA 111:12734–12739
Weigel P, Marc F, Simon S, Sakanyan V (2002) Ornithine N-acetyltransferase and arginine biosynthesis in thermophilic bacteria. In: Recent Research Developments in Microbiology 6:95-106. Research Signpost. S.G. Pandalai (editor)
Wintjens R, Liévin J, Rooman M, Buisine E (2000) Contribution of cation–π interactions to the stability of protein-DNA complexes. J Mol Biol 302:395–410
Wissenbach U, Keck B, Unden G (1993) Physical map location of the new artPIQMJ genes of Escherichia coli, encoding a periplasmic arginine transport system. J Bacteriol 175:3687–3688
Wissenbach U, Six S, Bongaerts J, Ternes D, Steinwachs S, Unden G (1995) A third periplasmic transport system for l-arginine in Escherichia coli: molecular characterization of the artPIQMJ genes, arginine binding and transport. Mol Microbiol 17:675–686
Wu WH, Morris DR (1973) Biosynthetic arginine decarboxylase from Escherichia coli. Purification and properties. J Biol Chem 248:1687–1695
Xu Y, Liang Z, Legrain C, Ruger H, Glansdorff N (2000) Evolution of arginine biosynthesis in the bacterial domain: novel gene-enzyme relationships from psychrophilic Moritella strains (Vibrionaceae) and evolutionary significance of N-α-acetylornithinase. J Bacteriol 182:1609–1615
Xu Y, Glansdorff N, Labedan B (2006) Bioinformatic analysis of an unusual gene-enzyme relationship in the arginine biosynthetic pathway among marine gamma proteobacteria: implications concerning the formation of N-acetylated intermediates in prokaryotes. BMC Genom 7:4. https://doi.org/10.1184/1471-2164-7-4
Xu Y, Labedan B, Glansdorff N (2007) Surprising arginine biosynthesis: a reappraisal of the enzymology and evolution of the pathway. Microbiol Mol Biol Rev 71:36–47
York MK, Stodolski M (1981) Characterization of P1 arg derivatives from Escherichia coli K-12 transduction. I. IS1 elements flank the argF gene segment. Mol Gen Genet 181:230–240
Zhao B, Houry WA (2010) Acid stress response in enteropathogenic gammaproteobacteria: an aptitude for survival. Biochem Cell Biol 88:301–314
Zhou X, Lou Z, Fu S, Yang A, Shen H, Li Z, Feng Y, Bartlam M, Wang H, Rao Z (2010) Crystal structure of ArgP from Mycobacterium tuberculosis confirms two distinct conformations of full-length LysR transcriptional regulators and reveals its function in DNA binding and transcriptional regulation. J Mol Biol 396:1012–1024
Zimmer DP, Soupene E, Lee HL, Wendisch VF, Khodursky AB, Peter BJ, Bender RA, Kustu S (2000) Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc Natl Acad Sci USA 97:14674–14679
Zúñiga M, Pérez G, González-Candelas (2002) Evolution of arginine deiminase (ADI) pathway genes. Mol Phylogenet Evol 25:429–444
Zwir I, Shin D, Kato A, Nishino K, Latifi T, Solomon F, Hare JM, Huang H, Groisman EA (2005) Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci USA 102:2862–2867
Acknowledgements
Work performed in our laboratory was supported by research grants from the Fonds Wetenschappelijk Onderzoek-Vlaanderen (Research Foundation-Flanders G.0056.03, G.0014.02N, G.0429.06, G.0321.13N) and the Research Council of the Vrije Universiteit Brussel. This review is dedicated to the memory of the late Nicolas Glansdorff, outstanding microbiologist and lecturer, devoted pioneer of arginine metabolism, with a passion for the origin and evolution of life.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants for whom identifying information is included in this article.
Additional information
Handling Editor: J. D. Wade.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Charlier, D., Bervoets, I. Regulation of arginine biosynthesis, catabolism and transport in Escherichia coli. Amino Acids 51, 1103–1127 (2019). https://doi.org/10.1007/s00726-019-02757-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-019-02757-8