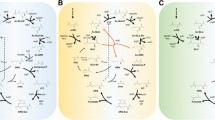
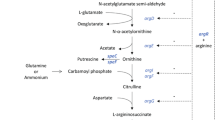
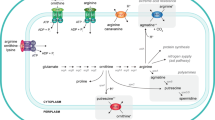
Abstract
l-Arginine is produced by bacterial fermentation and is consumed in food flavoring and pharmaceutical industries. A better understanding of arginine metabolism in bacteria could be beneficial for a rational design of recombinant l-arginine producers by genetic engineering. This mini-review illustrated the current status of genes and enzymes for arginine metabolism, including biosynthetic pathways, catabolic pathways, uptake and excretion systems, and regulation. The linkage of polyamine and glutamate metabolism to the arginine network was also discussed, followed by a perspective view on how to construct arginine overproducing strains of bacteria with increasing biosynthesis and excretion and decreasing catabolism and uptake.


Similar content being viewed by others
References
Abdelal A (1979) Arginine catabolism by microorganisms. Annu Rev Microbiol 33:139–168
Ali NO, Jeusset J, Larquet E, Le Cam E, Belitsky B, Sonenshein AL, Msadek T, Debarbouille M (2003) Specificity of the interaction of RocR with the rocG–rocA intergenic region in Bacillus subtilis. Microbiology 149:739–750
Barcelona-Andres B, Marina A, Rubio V (2002) Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J Bacteriol 184:6289–6300
Belitsky BR (2004) Bacillus subtilis GabR, a protein with DNA-binding and aminotransferase domains, is a PLP-dependent transcriptional regulator. J Mol Biol 340:655–664
Belitsky BR, Kim HJ, Sonenshein AL (2004) CcpA-dependent regulation of Bacillus subtilis glutamate dehydrogenase gene expression. J Bacteriol 186:3392–3398
Belitsky BR, Sonenshein AL (1998) Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J Bacteriol 180:6298–6305
Belitsky BR, Sonenshein AL (1999) An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc Natl Acad Sci U S A 96:10290–10295
Belitsky BR, Sonenshein AL (2002) GabR, a member of a novel protein family, regulates the utilization of gamma-aminobutyrate in Bacillus subtilis. Mol Microbiol 45:569–583
Bellmann A, Vrljic M, Patek M, Sahm H, Kramer R, Eggeling L (2001) Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology 147:1765–1774
Bockholt R, Scholten-Beck G, Pistorius EK (1996) Construction and partial characterization of an l-amino acid oxidase-free Synechococcus PCC 7942 mutant and localization of the l-amino acid oxidase in the corresponding wild type. Biochim Biophys Acta 1307:111–121
Bourdineaud JP, Heierli D, Gamper M, Verhoogt HJ, Driessen AJ, Konings WN, Lazdunski C, Haas D (1993) Characterization of the arcD arginine:ornithine exchanger of Pseudomonas aeruginosa. Localization in the cytoplasmic membrane and a topological model. J Biol Chem 268:5417–5424
Burkovski A, Kramer R (2002) Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Appl Microbiol Biotechnol 58:265–274
Calogero S, Gardan R, Glaser P, Schweizer J, Rapoport G, Debarbouille M (1994) RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J Bacteriol 176:1234–1241
Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW (1999) Control of acid resistance in Escherichia coli. J Bacteriol 181:3525–3535
Celis RT (1999) Repression and activation of arginine transport genes in Escherichia coli K 12 by the ArgP protein. J Mol Biol 294:1087–1095
Cho K, Fuqua C, Martin BS, Winans SC (1996) Identification of Agrobacterium tumefaciens genes that direct the complete catabolism of octopine. J Bacteriol 178:1872–1880
Cho K, Fuqua C, Winans SC (1997) Transcriptional regulation and locations of Agrobacterium tumefaciens genes required for complete catabolism of octopine. J Bacteriol 179:1–8
Cunin R, Glansdorff N, Pierard A, Stalon V (1986) Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev 50:314–352
Dennis CC, Glykos NM, Parsons MR, Phillips SE (2002) The structure of AhrC, the arginine repressor/activator protein from Bacillus subtilis. Acta Crystallogr D Biol Crystallogr 58:421–430
Dessaux Y, Petit A, Tempe J, Demarez M, Legrain C, Wiame JM (1986) Arginine catabolism in Agrobacterium strains: role of the Ti plasmid. J Bacteriol 166:44–50
Devroede N, Thia-Toong TL, Gigot D, Maes D, Charlier D (2004) Purine and pyrimidine-specific repression of the Escherichia coli carAB operon are functionally and structurally coupled. J Mol Biol 336:25–42
D'Hooghe I, Vander Wauven C, Michiels J, Tricot C, de Wilde P, Vanderleyden J, Stalon V (1997) The arginine deiminase pathway in Rhizobium etli: DNA sequence analysis and functional study of the arcABC genes. J Bacteriol 179:7403–7409
Fan CL, Rodwell VW (1975) Physiological consequences of starvation in Pseudomonas putida: degradation of intracellular protein and loss of activity of the inducible enzymes of l-arginine catabolism. J Bacteriol 124:1302–1311
Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL (1997) Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev 61:393–410
Gardan R, Rapoport G, Debarbouille M (1995) Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J Mol Biol 249:843–856
Glansdorff N (1996) Biosynthesis of arginine and polyamines. In: Neidhardt FC (ed) Escherichia coli and Salmonella: cellular and molecular biology, 2nd edn. ASM, Washington, DC, pp 408–433
Gobert AP, McGee DJ, Akhtar M, Mendz GL, Newton JC, Cheng Y, Mobley HL, Wilson KT (2001) Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc Natl Acad Sci U S A 98:13844–13849
Gong S, Richard H, Foster JW (2003) YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J Bacteriol 185:4402–4409
Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI (1998) PmrA–PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol 27:1171–1182
Haas D, Galimand M, Gamper M, Zimmermann A (1990) Arginine network of Pseudomonas aeruginosa: specific and global controls. In: Silver S, Chakraberty AM, Iglewski B, Kaplan S (eds) Pseudomonas: biotransformations, pathogenesis, and evolving biotechnology. ASM Press, Washington DC, pp 303–316
Han X, Turnbough CL Jr (1998) Regulation of carAB expression in Escherichia coli occurs in part through UTP-sensitive reiterative transcription. J Bacteriol 180:705–713
Hashim S, Kwon DH, Abdelal A, Lu CD (2004) The arginine regulatory protein mediates repression by arginine of the operons encoding glutamate synthase and anabolic glutamate dehydrogenase in Pseudomonas aeruginosa. J Bacteriol 186:3848–3854
Hegazy M (2004) Characterization of the arginine decarboxylase pathway in Pseudomonas aeruginosa PAO1. Ph.D. dissertation, Biology Department, Georgia State University, Atlanta, p 90
Higgins CF, Ames GF (1981) Two periplasmic transport proteins which interact with a common membrane receptor show extensive homology: complete nucleotide sequences. Proc Natl Acad Sci U S A 78:6038–6042
Ikeda M (2003) Amino acid production processes. Adv Biochem Eng Biotechnol 79:1–35
Itoh Y (1997) Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa. J Bacteriol 179:7280–7290
Itoh Y, Nakada Y (2004) Arginine and polyamine metabolism. In: Ramos J (ed) Pseudomonas. Kluwer/Plenum, New York, pp 243–272
Jann A, Matsumoto H, Haas D (1988) The fourth arginine catabolic pathway of Pseudomonas aeruginosa. J Gen Microbiol 134:1043–1053
Kiupakis AK, Reitzer L (2002) ArgR-independent induction and ArgR-dependent superinduction of the astCADBE operon in Escherichia coli. J Bacteriol 184:2940–2950
Krin E, Laurent-Winter C, Bertin PN, Danchin A, Kolb A (2003) Transcription regulation coupling of the divergent argG and metY promoters in Escherichia coli K-12. J Bacteriol 185:3139–3146
Kurihara S, Oda S, Kato K, Kim HG, Koyanagi T, Kumagai H, Suzuki H (2005) A novel putrescine utilization pathway involves gamma-glutamylated intermediates of Escherichia coli K-12. J Biol Chem 280:4602–4608
Kwon DH, Lu CD, Walthall DA, Brown TM, Houghton JE, Abdelal AT (1994) Structure and regulation of the carAB operon in Pseudomonas aeruginosa and Pseudomonas stutzeri: no untranslated region exists. J Bacteriol 176:2532–2542
Larsen R, Kok J, Kuipers OP (2005) Interaction between ArgR and AhrC controls regulation of arginine metabolism in Lactococcus lactis. J Biol Chem 280:19319–19330
Lu CD, Abdelal AT (1999) Role of ArgR in activation of the ast operon, encoding enzymes of the arginine succinyltransferase pathway in Salmonella typhimurium. J Bacteriol 181:1934–1938
Lu CD, Abdelal AT (2001) The gdhB gene of Pseudomonas aeruginosa encodes an arginine-inducible NAD(+)-dependent glutamate dehydrogenase which is subject to allosteric regulation. J Bacteriol 183:490–499
Lu CD, Itoh Y, Nakada Y, Jiang Y (2002) Functional analysis and regulation of the divergent spuABCDEFGH–spuI operons for polyamine uptake and utilization in Pseudomonas aeruginosa PAO1. J Bacteriol 184:3765–3773
Lu CD, Winteler H, Abdelal A, Haas D (1999) The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J Bacteriol 181:2459–2464
Lu CD, Yang Z, Li W (2004) Transcriptome analysis of the ArgR regulon in Pseudomonas aeruginosa. J Bacteriol 186:3855–3861
Luthi E, Baur H, Gamper M, Brunner F, Villeval D, Mercenier A, Haas D (1990) The arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa contains an additional gene, arcD, encoding a membrane protein. Gene 87:37–43
Macfarlane EL, Kwasnicka A, Hancock RE (2000) Role of Pseudomonas aeruginosa PhoP–PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146:2543–2554
Maghnouj A, Abu-Bakr AA, Baumberg S, Stalon V, Vander Wauven C (2000) Regulation of anaerobic arginine catabolism in Bacillus licheniformis by a protein of the Crp/Fnr family. FEMS Microbiol Lett 191:227–234
Maghnouj A, de Sousa Cabral TF, Stalon V, Vander Wauven C (1998) The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor ArgR. J Bacteriol 180:6468–6475
Makarova KS, Mironov AA, Gelfand MS (2001) Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol 2:RESEARCH0013
Martin RG, Rosner JL (2001) The AraC transcriptional activators. Curr Opin Microbiol 4:132–137
Morris SMJ, Loscalzo J, Bier D, Souba WW (eds) (2004) Arginine metabolism: enzymology, nutrition, and clinical significance. The Journal of Nutrition, Bermuda
Nakada Y, Itoh Y (2002) Characterization and regulation of the gbuA gene, encoding guanidinobutyrase in the arginine dehydrogenase pathway of Pseudomonas aeruginosa PAO1. J Bacteriol 184:3377–3384
Nakada Y, Itoh Y (2003) Identification of the putrescine biosynthetic genes in Pseudomonas aeruginosa and characterization of agmatine deiminase and N-carbamoylputrescine amidohydrolase of the arginine decarboxylase pathway. Microbiology 149:707–714
Nakada Y, Jiang Y, Nishijyo T, Itoh Y, Lu CD (2001) Molecular characterization and regulation of the aguBA operon, responsible for agmatine utilization in Pseudomonas aeruginosa PAO1. J Bacteriol 183:6517–6524
Nandineni MR, Gowrishankar J (2004) Evidence for an arginine exporter encoded by yggA (argO) that is regulated by the LysR-type transcriptional regulator ArgP in Escherichia coli. J Bacteriol 186:3539–3546
Naumoff DG, Xu Y, Glansdorff N, Labedan B (2004) Retrieving sequences of enzymes experimentally characterized but erroneously annotated: the case of the putrescine carbamoyltransferase. BMC Genomics 5:52
Ni J, Sakanyan V, Charlier D, Glansdorff N, Van Duyne GD (1999) Structure of the arginine repressor from Bacillus stearothermophilus. Nat Struct Biol 6:427–432
Nishijyo T, Haas D, Itoh Y (2001) The CbrA–CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol Microbiol 40:917–931
Nishijyo T, Park SM, Lu CD, Itoh Y, Abdelal AT (1998) Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J Bacteriol 180:5559–5566
Ohtani K, Bando M, Swe T, Banu S, Oe M, Hayashi H, Shimizu T (1997) Collagenase gene (colA) is located in the 3′-flanking region of the perfringolysin O (pfoA) locus in Clostridium perfringens. FEMS Microbiol Lett 146:155–159
Park SM, Lu CD, Abdelal AT (1997a) Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J Bacteriol 179:5300–5308
Park SM, Lu CD, Abdelal AT (1997b) Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons. J Bacteriol 179:5309–5317
Quinn CL, Stephenson BT, Switzer RL (1991) Functional organization and nucleotide sequence of the Bacillus subtilis pyrimidine biosynthetic operon. J Biol Chem 266:9113–9127
Quintero MJ, Muro-Pastor AM, Herrero A, Flores E (2000) Arginine catabolism in the cyanobacterium Synechocystis sp. Strain PCC 6803 involves the urea cycle and arginase pathway. J Bacteriol 182:1008–1015
Rajagopal BS, DePonte J, Tuchman M, Malamy MH (1998) Use of inducible feedback-resistant N-acetylglutamate synthetase (argA) genes for enhanced arginine biosynthesis by genetically engineered Escherichia coli K-12 strains. Appl Environ Microbiol 64:1805–1811
Reitzer L (1996) Sources of nitrogen and their utilization. In: Neidhardt FC (ed) Escherichia coli and Salmonella. ASM Press, Washington, DC, pp 380–407
Ruepp A, Soppa J (1996) Fermentative arginine degradation in Halobacterium salinarium (formerly Halobacterium halobium): genes, gene products, and transcripts of the arcRACB gene cluster. J Bacteriol 178:4942–4947
Sanchez R, Roovers M, Glansdorff N (2000) Organization and expression of a Thermus thermophilus arginine cluster: presence of unidentified open reading frames and absence of a Shine-Dalgarno sequence. J Bacteriol 182:5911–5915
Schneider BL, Kiupakis AK, Reitzer LJ (1998) Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J Bacteriol 180:4278–4286
Schneider BL, Ruback S, Kiupakis AK, Kasbarian H, Pybus C, Reitzer L (2002) The Escherichia coli gabDTPC operon: specific gamma-aminobutyrate catabolism and nonspecific induction. J Bacteriol 184:6976–6986
Shi D, Gallegos R, DePonte J, Morizono H, Yu X, Allewell NM, Malamy M, Tuchman M (2002) Crystal structure of a transcarbamylase-like protein from the anaerobic bacterium Bacteroides fagilis at 2.0 A resolution. J Biol Chem 320:899–908
Shi D, Morizono H, Yu X, Roth L, Caldovic L, Allewell NM, Malamy MH, Tuchman M (2005) Crystal structure of N-acetylornithine transcarbamylase from Xanthomonas campestris: a novel enzyme in a new arginine biosynthetic pathway found in several eubacteria. J Biol Chem 280:14366–14369
Stalon V, Vander Wauven C, Momin P, Legrain C (1987) Catabolism of arginine, citrulline and ornithine by Pseudomonas and related bacteria. J Gen Microbiol 133:2487–2495
Stim-Herndon KP, Flores TM, Bennett GN (1996) Molecular characterization of adiY, a regulatory gene which affects expression of the biodegradative acid-induced arginine decarboxylase gene (adiA) of Escherichia coli. Microbiology 142:1311–1320
Szumanski MB, Boyle SM (1990) Analysis and sequence of the speB gene encoding agmatine ureohydrolase, a putrescine biosynthetic enzyme in Escherichia coli. J Bacteriol 172:538–547
Tabor CW, Tabor H (1985) Polyamines in microorganisms. Microbiol Rev 49:81–99
Thoden JB, Holden HM, Wesenberg G, Roaushel FM, Rayment I (1997) Structure of carbamoylphosphate synthetase: a journal of 96 A from substrate to product. Biochemistry 36:6305–6316
Tonon T, Bourdineaud JP, Lonvaud-Funel A (2001) The arcABC gene cluster encoding the arginine deiminase pathway of Oenococcus oeni, and arginine induction of a CRP-like gene. Res Microbiol 152:653–661
Tricot C, Stalon V, Legrain C (1991) Isolation and characterization of Pseudomonas putida mutants affected in arginine, ornithine and citrulline catabolism: function of the arginine oxidase and arginine succinyltransferase pathways. J Gen Microbiol 137:2911–2918
Tricot C, Vander Wauven C, Wattiez R, Falmagne P, Stalon V (1994) Purification and properties of a succinyltransferase from Pseudomonas aeruginosa specific for both arginine and ornithine. Eur J Biochem 224:853–861
Tuchman M, Rajagopal BS, McCann MT, Malamy MH (1997) Enhanced production of arginine and urea by genetically engineered Escherichia coli K-12 strains. Appl Environ Microbiol 63:33–38
Utagawa T (2004) Production of arginine by fermentation. J Nutr 134:2854S–2867
Van Duyne GD, Ghosh G, Maas WK, Sigler PB (1996) Structure of the oligomerization and l-arginine binding domain of the arginine repressor of Escherichia coli. J Mol Biol 256:377–391
Vander Wauven C, Stalon V (1985) Occurrence of succinyl derivatives in the catabolism of arginine in Pseudomonas cepacia. J Bacteriol 164:882–886
Vanderbilt AS, Gaby NS, Rodwell VW (1975) Intermediates and enzymes between alpha-ketoarginine and gamma-guanidinobutyrate in the l-arginine catabolic pathway of Pseudomonas putida. J Biol Chem 250:5322–5329
Verhoogt HJ, Smit H, Abee T, Gamper M, Driessen AJ, Haas D, Konings WN (1992) arcD, the first gene of the arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa, encodes an arginine–ornithine exchanger. J Bacteriol 174:1568–1573
Vrljic M, Garg J, Bellmann A, Wachi S, Freudl R, Malecki MJ, Sahm H, Kozina VJ, Eggeling L, Saier MH (1999) The LysE superfamily: topology of the lysine exporter LysE of Corynebacterium glutamicum, a paradigm for a novel superfamily of transmembrane solute translocators. J Mol Microbiol Biotechnol 1:327–336
Wissenbach U, Six S, Bongaerts J, Ternes D, Steinwachs S, Unden G (1995) A third periplasmic transport system for l-arginine in Escherichia coli: molecular characterization of the artPIQMJ genes, arginine binding and transport. Mol Microbiol 17:675–686
Yang H, Park SM, Nolan WG, Lu CD, Abdelal AT (1997) Cloning and characterization of the arginine-specific carbamoyl-phosphate synthetase from Bacillus stearothermophilus. Eur J Biochem 249:443–449
Zuniga M, Miralles Md, Mdel C, Perez-Martinez G (2002) The product of arcR, the sixth gene of the arc operon of Lactobacillus sakei, is essential for expression of the arginine deiminase pathway. Appl Environ Microbiol 68:6051–6058
Acknowledgements
The author is grateful to John E. Houghton for critical reading and to Zhe Yang for the excellent artwork of the manuscript. Research in the author's laboratory was supported by the National Science Foundation grant no. 0415608.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lu, CD. Pathways and regulation of bacterial arginine metabolism and perspectives for obtaining arginine overproducing strains. Appl Microbiol Biotechnol 70, 261–272 (2006). https://doi.org/10.1007/s00253-005-0308-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-005-0308-z