Abstract
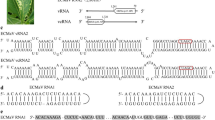
A new positive-strand RNA virus genome was discovered in Camellia japonica plants. The complete genome of the virus is 12,570 nt in size, excluding the poly(A) tail, and contains one large open reading frame (ORF1) and two small open reading frames (ORF2, ORF3). ORF1 and ORF2 are homologous to sequences of waikaviruses, while ORF3 has no relatives in the databases. ORF1 encodes a putative polyprotein precursor that is putatively processed into eight smaller proteins, as in typical waikaviruses. Comprehensive analysis, including BLAST searches, genome organization and pairwise sequence comparisons, and phylogeny reconstructions, invariably placed the virus with the waikaviruses. Furthermore, due to lower amino acid sequence identity to known waikaviruses than the threshold species demarcation cutoff, this virus may represent a new species in the genus Waikavirus, family Secoviridae, and we have tentatively named it “camellia virus A” (CamVA). Finally, a field survey was conducted to assess the occurrence of CamVA in camellias and its associated symptoms.


Similar content being viewed by others
References
Thompson JR, Dasgupta I, Fuchs M, Iwanami T, Karasev AV, Petrzik K, Sanfaçon H, Tzanetakis I, van der Vlugt R, Wetzel T, Yoshikawa N (2017) ICTV virus taxonomy profile: Secoviridae. J Gen Virol 98:529–531
Wylie SJ, Luo H, Li H, Jones MGK (2011) Multiple polyadenylated RNA viruses detected in pooled cultivated and wild plant samples. Arch Virol 157:271–284
Rott ME, Tremaine JH, Rochon DM (1991) Nucleotide sequence of tomato ringspot virus RNA-2. J Gen Virol 72:1505–1514
Wetzel T, Beck A, Wegener U, Krczal G (2004) Complete nucleotide sequence of the RNA 1 of a grapevine isolate of arabis mosaic virus. Arch Virol 149:989–995
Sailaja B, Anjum N, Patil YK, Agarwal S, Malathi P, Krishnaveni D, Balachandran SM, Viraktamath BC, Mangrauthia SK (2013) The complete genome sequence of a south Indian isolate of rice tungro spherical virus reveals evidence of genetic recombination between distinct isolates. Virus Genes 47:515–523
Murant AF, Roberts IM, Elnagar S (1976) Association of virus-like particles with the foregut of the aphid Cavariella aegopodii transmitting the semi-persistent viruses anthriscus yellows and parsnip yellow fleck. J Gen Virol 31:47–57
Seo JK, Kwak HR, Lee Y, Kim J, Kim MK, Kim CS, Choi HS (2015) Complete genome sequence of bellflower vein chlorosis virus, a novel putative member of the genus Waikavirus. Arch Virol 160:3139–3142
Reddick BB, Habera LF, Law MD (1997) Nucleotide sequence and taxonomy of maize chlorotic dwarf virus within the family Sequiviridae. J Gen Virol 78:1165–1174
Sebastian L (1996) Molecular mapping of resistance to rice tungro spherical virus and green leafhopper. Phytopathology 86:25
Wayadande AC, Nault LR (1993) Leafhopper probing behavior associated with maize chlorotic dwarf virus transmission to maize. Phytopathology 83:522
Wu Y, Ning Z, Tian B, Wang Y, Zhen L (2015) Origin and differences of camellia between China and Japan. World For Res 28:81–84
Zhang S, Shen P, Li M, Tian X, Zhou C, Cao M (2018) Discovery of a novel geminivirus associated with camellia chlorotic dwarf disease. Arch Virol 163:1709–1712
Zhang S, Yang L, Ma L, Tian X, Li R, Zhou C, Cao M (2020) Virome of Camellia japonica: discovery of and molecular characterization of new viruses of different taxa in camellias. Front Microbiol 11:945
Liu H, Wu L, Zheng L, Cao M, Li R (2018) Characterization of three new viruses of the family Betaflexiviridae associated with camellia ringspot disease. Virus Res 272:197668
Hao X, Zhang W, Zhao F, Liu Y, Qian W, Wang Y, Wang L, Zeng J, Yang Y, Wang X (2018) Discovery of plant viruses from tea plant (Camellia sinensis (L.) O. Kuntze) by metagenomic sequencing. Front Microbiol 9:2175
Wu L, Du M, Xiao H, Peng L, Li R (2020) Complete genomic sequence of tea-oil camellia deltapartitivirus 1, a novel virus from Camellia oleifera. Arch Virol 165:227–231
Zheng L, Chen M, Li R (2020) Camellia ringspot-associated virus 4, a proposed new foveavirus from Camellia japonica. Arch Virol 165:1707–1710
Peracchioa C, Forgiaab M, Chiapelloa M, Vallinoa M, Turinaa M, Ciuffo M (2020) A complex virome including two distinct emaraviruses associated with virus-like symptoms in Camellia japonica. Virus Res 286:197964
Li R, Zheng L, Cao M, Wu L, Liu H (2020) First identification and molecular characterization of a new badnavirus infecting camellia. Arch Virol 165:2115–2118
Standley D (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Sudhir K, Glen S, Li M, Christina K, Koichiro T (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Koloniuk I, Fránová J (2018) Complete nucleotide sequence and genome organization of red clover associated virus 1 (RCaV1), a putative member of the genus Waikavirus (family Secoviridae, order Picornavirales). Arch Virol 163:3447–3449
Park D, Hahn Y (2019) A novel Waikavirus (the family Secoviridae) genome sequence identified in rapeseed (Brassica napus). Acta Virol 63:211–216
Thekke-Veetil T, Ho T, Postman JD, Tzanetakis IE (2020) Blackcurrant waikavirus A, a new member of the genus Waikavirus, and its phylogenetic and molecular relationship with other known members. Eur J Plant Pathol 157:59–64
Acknowledgements
This research was supported by the National Natural Science Foundation of China (32072389), Innovation Program for Chongqing’s Overseas Returnees (cx2019013), and the 111 Project (B18044).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Compliance with ethical standards
I have read and abided by the statement of ethical standards for manuscripts submitted to Archives of Virology.
Conflict of interest
All authors declare they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Sead Sabanadzovic.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liao, R., Chen, Q., Zhang, S. et al. Complete genome sequence of camellia virus A, a tentative new member of the genus Waikavirus. Arch Virol 166, 3207–3210 (2021). https://doi.org/10.1007/s00705-021-05216-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05216-6