Abstract
Pigs are susceptible to infection with both human and avian influenza A viruses and are considered intermediate hosts that facilitate virus reassortment. Although H5N1 virus has spread to a wide range of avian and mammalian species, data about swine H5N1 isolates are scarce. To determine whether Asian H5N1 influenza viruses had been transmitted to pigs, a total of 1,107 nasal swab samples from healthy swine were collected from 2008 to 2009 in Jiangsu province of eastern China. In this survey, two H5N1 viruses A/swine/Jiangsu/1/2008 (JS/08) and A/swine/Jiangsu/2/2009 (JS/09) were isolated and identified. Phylogenetic analysis showed that JS/08 and JS/09 belonged to clade 7 and clade 2.3.4, respectively, and shared over 99.0 % sequence identity with poultry H5N1 isolates of the same clade in China. Receptor specificity analysis also showed that both of the swine H5N1 isolates bound preferentially to avian-type receptors. However, experiments in mammals indicated that JS/09 was moderately pathogenic to mice without prior adaption, whereas JS/08 had limited ability to replicate. Our findings suggest that pigs are naturally infected with avian H5N1 virus and highlight the potential threat to public health due to adaption or reassortment of H5N1 virus in this species.
Similar content being viewed by others
Introduction
In 1996, the Asian highly pathogenic avian influenza (HPAI) H5N1 virus was initially recognized in geese in Guangdong Province of China, and the first case of human infection was reported in Hong Kong in 1997 [6, 32, 36]. Since then, Asian H5N1 viruses present a continuing threat to both animals and public health on a global scale [27, 38]. Although a number of avian H5N1 outbreaks have been reported, data about H5N1 in pigs are scarce. It has been shown that pigs have limited susceptibility to infection with avian H5N1 viruses under experimental conditions [5, 20, 37]. However, serological surveillance revealed evidence of H5N1 infection in swine in Vietnam and co-circulation with other subtypes of avian influenza virus in China [5, 22]. The isolation of H5N1 viruses from pigs has also been reported in China and Indonesia [25, 30, 42].
Pigs are considered intermediate hosts or “mixing vessels” because their tracheal epithelial cells express both SAα2,3Gal and SAα2,6Gal receptors [3, 12, 14]. This is the molecular basis for pigs being susceptible to infection with both avian and human influenza viruses, which facilitates virus reassortment and can potentially lead to pandemics, as evidenced by the occurrence of the 2009 H1N1 pandemic virus [8, 31, 34]. Since pigs are naturally infected with H5N1, the potential risk of H5N1 reassortment and introduction of novel deadly viruses back into the human population exists. Recent studies have demonstrated the high genetic compatibility between avian H5N1 viruses and the currently circulating human influenza viruses [4, 16, 26]. Moreover, evidence from clinical surveillance has also shown that avian H5 subtype influenza A viruses participated in the reassortment of swine human-like H3N2 viruses isolated in China [2, 7]. Hence, understanding the prevalence and adaptation of H5N1 influenza viruses in the pig population is of crucial importance.
Although several HPAI H5N1 viruses have been isolated from pigs in Fujian and Shandong provinces of eastern China, the H5N1 situation of Jiangsu Province in the same area is still unknown [30, 42]. In this study, two swine H5N1 viruses isolated in Jiangsu Province were characterized genetically and pathobiologically, and their receptor specificity and pathogenicity were also investigated. Our results further demonstrate the importance of pigs as intermediate hosts for influenza A virus and provide useful information for improving surveillance in swine.
Materials and methods
Virus isolation and identification
The first period of our influenza virus survey in pigs in Jiangsu Province of China was performed from October 2008 to May 2009. In this survey, a total of 1,107 nasal swab samples were randomly collected from healthy swine. These samples were maintained in transport medium containing antibiotics and kept on ice until transported to the laboratory. After centrifugation for 10 min at 5000 rpm at 4 °C, 0.2 ml of the supernatant was inoculated into the allantoic cavities of 10-day-old specific-pathogen-free (SPF) embryonated chicken eggs. After 48-72 h of incubation at 35 °C, the allantoic fluids were harvested and tested for hemagglutinin (HA) activity. The HA subtypes and neuraminidase (NA) subtypes of positive isolates were identified by sequence analysis of the reverse transcription polymerase chain reaction (RT-PCR) products as described previously [15, 28]. All of the experiments dealing with H5N1 viruses were conducted in biosafety level 3 facilities. The animal research was approved by the Jiangsu Administrative Committee for Laboratory Animals (Permit number: SYXKSU-2007-0005) and complied with the guidelines of Jiangsu Laboratory Animal Welfare and Ethical of Jiangsu Administrative Committee of Laboratory Animals.
Sequencing and phylogenetic analysis
Viral RNA was extracted from infected allantoic fluid using TRIzol LS Reagent (Invitrogen) and reverse transcribed with the Uni12 primer (5’-AGCAAAAGCAGG-3’). PCR was performed using specific primers as described [10]. Each full-length PCR product was purified and sequenced. The sequence data were compiled and edited using DNASTAR, Lasergene v7.1. To assess the genetic relationships among H5 influenza virus strains, reference gene sequences were selected from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), and a phylogenetic tree was constructed by the neighbor-joining method using MEGA 4.1 software. Bootstrap values were calculated based on 1000 replicates of the alignment.
Hemagglutination assay for receptor-binding analysis
To measure if the H5N1 viruses had adapted to recognize human-type receptors during replication in pigs, we performed a hemagglutination assay as described previously [33]. Briefly, a 100-μl aliquot of a 10 % suspension of goose red blood cells (GRBCs) was treated with 1.25 units of α2, 3-sialidase (TaKaRa) for 1 h at 37 °C. The treated GRBCs were washed twice and adjusted to a final working concentration of 0.5 % with phosphate-buffered saline (PBS). Viruses (50 μl) were serially diluted with 50 μl of PBS and mixed with 50 μl of 0.5 % of GRBCs in a 96-well microtiter plate. The reaction was kept at room temperature, and HA titers were read after a 15-minute incubation. The A(H1N1)pdm2009 virus A/California/04/2009 (CA/09) and poultry H5N1 isolate A/mallard/Huadong/S/2005 (HD/05) were used as controls.
Solid-phase direct binding assay
The receptor specificity was further analyzed by a solid-phase direct binding assay as described previously [1]. Briefly, the synthetic sialylglycopolymers Neu5Aca2-3Galb1-4GlcNAcb (3’SLN)-PAA-biotin and Neu5Aca2-3Galb1-4GlcNAcb (6’SLN)-PAA-biotin (GlycoTech) were serially diluted in PBS and added to the wells of 96-well streptavidin coated microtiter plates (Pierce). The plates were blocked with PBS containing 2 % skim milk powder, and 128 HA units of live virus was added per well. Chicken antiserum against the virus was diluted in PBS and added to each well. Bound antibody was detected by sequential addition of HRP-conjugated rabbit anti-chicken IgG antibody and tetramethylbenzidine substrate solution. The reaction was stopped with 1 M H2SO4, and the absorbance at 450 nm was read. Each sample was measured in triplicate.
Replication kinetics
Madin-Darby canine kidney (MDCK) cells were infected with 0.01 50 % tissue culture infective dose (TCID50) of virus for 1 h at 37 °C. Cells were washed twice with PBS and incubated in minimum essential medium at time zero. Aliquots of culture supernatants were collected at 12, 24, 48 and 72 hours post-inoculation (h.p.i) and immediately stored at -80 °C for determination of virus titers. Virus was titrated (log10 TCID50) in MDCK cells by the method of Reed and Muench [29].
Plaque assays
Confluent MDCK cell monolayers in six-well plates were infected with serial tenfold dilutions of virus for 1 h at 37 °C. Cells were washed twice with PBS and covered with 0.8 % agarose in 2× Dulbecco’s Modified Eagle Medium (DMEM) containing 0.3 % BSA and 1 μg/ml of TPCK-trypsin (Sigma, Missouri, USA). After 48 h of incubation at 37 °C, the agarose was removed and the cells were fixed with 10 % formaldehyde and then stained with 0.1 % crystal violet. The plaques produced by the viruses were measured using the GNU image manipulation program, version 2.8 (www.gimp.org), as described previously [24].
Mouse study
To determine the 50 % mouse lethal dose (MLD50), two groups of twenty 6-week-old female BALB/c mice were infected intranasally with 103 to 106 50 % egg infective dose (EID50) of the JS/08 and JS/09 viruses diluted in 50 μl PBS. The MLD50 values were calculated by the method of Reed and Muench after a 14-day observation period and expressed as EID50. To determine the morbidity and mortality, three groups of ten 6-week-old female BALB/c mice were infected intranasally with 106 EID50 of JS/08 and JS/09 viruses or mock inoculated with PBS in 50-μl volumes. Body weight was recorded daily until 14 days post-inoculation (d.p.i.). Infected mice showing more than 25 % body weight loss were humanely euthanized and recorded as dead.
To evaluate the replication ability of the JS/08 and JS/09 viruses in vivo, two groups of six mice were inoculated with 106 EID50 of the indicated viruses, and three mice from each infected group were euthanized humanely at 3 d.p.i. and 5 d.p.i. The lung, heart, brain, spleen, kidney and liver were collected and homogenized in PBS at a ratio of 1:1 (g/ml). After centrifugation for 30 min at 8,000 rpm, the supernatants were collected for the extraction of viral RNA as described above. The viral RNA was reverse transcribed to cDNA using random primers (Invitrogen).
Real-time PCR for quantitation of viral loads
Viral loads in each of the organ samples were determined by quantitative PCR. cDNA from tissues mentioned above were run in an ABI 7300 Real Time PCR System using an SYBR Premix Ex Taq Kit (TaKaRa). The detection targeting the M gene was performed with the forward primer 5’-TGGGATTTGTATTCACGCTCA-3’ and the reverse primer 5’-AGTAGCTGAGTGCGACCTCCTT-3’. A reference standard was prepared using pGEM-T Easy Vector (Promega) containing the corresponding target virus sequences. A series of tenfold dilutions equivalent to 1×102 to 1×108 copies per reaction were prepared to generate concomitant calibration curves. The detection limit of this assay was 820 copies per ml.
Statistical analysis
Statistical analysis was performed using PAWS Statistics 18 (SPSS Inc., USA). Significant differences in virus titers and plaque size between the two isolates were analyzed by an independent-sample t-test, and a P-value of 0.05 or less was considered significant.
Nucleotide sequence accession numbers
The nucleotide sequences of the viral genomes of the two H5N1 isolates were submitted to GenBank and are available under accession numbers KC683516-KC683531.
Results
Isolation and identification of viruses
From October 2008 to May 2009, 1,107 nasal swab samples were randomly collected from apparently healthy pigs (4–6 months of age) in Jiangsu Province of China. A total of six strains of influenza viruses were isolated and sequenced (Table 1). Among these viruses, two strains of H5N1 viruses were identified and designated as A/swine/Jiangsu/1/2008 (JS/08) and A/swine/Jiangsu/2/2009 (JS/09), respectively. These two H5N1 viruses were isolated from different pig farms in Yangzhou. The isolation rate of H5N1 virus was 0.18 % in this surveillance.
Phylogenetic analysis
Phylogenetic analysis of the HA genes of JS/08 and JS/09 showed that these two swine H5N1 isolates belonged to clade 7 and clade 2.3.4, respectively (Fig. 1a). JS/08 was mostly closely related to chicken isolate A/chicken/Jiangsu/18/2008, and the identities of the HA gene between these two viruses were 99.2 % for the nucleotide sequence and 99.1 % for the amino acid sequence. Its HA segment was also closely related to that of A/Chicken/Huadong/4/2008, which was detected in Jiangsu Province [17]. JS/09 showed the highest homology to A/tree sparrow/Jiangsu/1/2008, and the nucleotide and amino acid homology of the HA genes between these two viruses were 99.4 % and 99.1 %, respectively. JS/09 also showed a close relationship to the human isolate A/Jiangsu/1/2007, which shared highest homology with A/tree sparrow/Jiangsu/1/2008 as well [21]. The NA gene and all the internal gene trees showed the same pattern of common origin that was seen for the HA gene tree (Fig. 1b-h). With the exception of the PB2 gene, JS/08 showed the highest homology to A/chicken/Jiangsu/18/2008, and the NA and the other internal genes were mostly closely related to those of A/Chicken/Huadong/4/2008. The percentages of nucleotide and amino acid sequence similarities among these genes ranged from 99.2 to 99.9 % and 97.8 to 100 %, respectively. The NA and all the internal genes of JS/09 also showed high homology to those of A/tree sparrow/Jiangsu/1/2008, except the PA and M genes, which were most closely related to those of A/Jiangsu/1/2007. The percentages of nucleotide and amino acid sequence similarities among these genes ranged from 99.1 to 99.8 % and 98.7 to 99.8 %, respectively. Overall, these results indicated that each H5N1 isolate evolved from a common source of the same clade in Jiangsu Province of China.
Phylogenetic trees for the HA (a), NA (b), PB2 (c), PB1 (d), PA (e), NP (f), M (g) and NS (h) genes of the H5N1 influenza viruses isolated from swine in Jiangsu, China. Trees were constructed for H5 isolates and reference strains from GenBank based on their open reading frame sequences. The sequences determined in the present study are indicated by black triangles. The trees were generated using the neighbour-joining method in MEGA 4.1 with 1,000 bootstrap trials performed to assign confidence to the grouping
Receptor-binding analysis
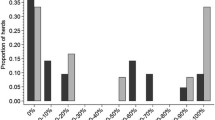
In the hemagglutination assay, compared to untreated GRBCs, the HD/05, JS/08 and JS/09 viruses showed no HA activity with α2,3-sialidase-treated GRBCs, which had only α2,6-receptors. However, full HA activity was detected with both types of GRBCs when the 2009 pandemic H1N1 virus CA/09 was tested (Table 2). To further evaluate the receptor-binding specificity, a solid-phase direct binding assay was conducted. The HD/05, JS/08 and JS/09 viruses still showed an absolute preference for α2,3-sialoglycan (3’SLN) binding, opposite to the absolute α2,6-sialoglycan (6’SLN) binding preference of CA/09 (Fig. 2). Neither the JS/08 nor the JS/09 virus acquired the ability to recognize a human-type receptor during its replication in pigs.
Solid-phase receptor-binding assay of human isolate A/California/04/2009 (a), poultry isolate A/mallard/Huadong/S/2005 (b), swine isolate A/swine/Jiangsu/1/2008 (c) and swine isolate A/swine/Jiangsu/2/2009 (d). Direct binding of viruses to sialylglycopolymers containing either 3’SLN-PAA or 6’SLN-PAA was measured. The data shown are representative of three independent binding experiments
Growth characteristics in vitro
The replication kinetics of the JS/08 and JS/09 viruses were studied in MDCK cells. At any given time point, JS/09 replicated to a higher titer than the JS/08 virus (Fig. 3a). The difference in the peak titer was noticable at 48 h.p.i., with a tenfold-higher titer for the JS/09 virus than for the JS/08 virus (P < 0.01). In the plaque assay, MDCK cells infected with JS/09 virus developed large plaques with a mean size of 2.58 ± 0.40 mm, whereas JS/08 formed significantly smaller plaques with an mean size of 1.32 ± 0.33 mm (P < 0.001) (Fig. 3b). These results indicated that JS/09 grew faster in MDCK cells and showed a more obvious cytopathic effect compared with JS/08.
Growth kinetics curves (a) and plaque assay (b) of JS/08 and JS/09 viruses in MDCK cells. Cells were infected at a multiplicity of infection of 0.01. Virus titers were determined in MDCK cells at the appropriate time points. Each data point on the curve indicates the mean ± standard deviation of three independent experiments. *, P < 0.05; **, P < 0.01. The sizes of plaques formed by the H5N1 viruses in MDCK cells at 48 hours postinfection are shown. The average plaque size of 10 representative plaques for each virus is indicated
Mouse study
To evaluate the virulence of JS/08 and JS/09 in mammals, we used a BALB/c mouse model to determine the MLD50. The results showed that JS/09 was more virulent for mice than JS/08; the MLD50 values of the two H5N1 isolates were 105.2 EID50 and greater than 106.0 EID50, respectively. To investigate their morbidity and mortality, mice were inoculated intranasally with 106.0 EID50 of JS/08 and JS/09. Approximately 25 % weight loss was detected in JS/09-infected mice, and the mortality rate was 100 %. The JS/09-infected mice also exhibited clinical signs, including depression, huddling and decreased activity. In contrast, JS/08-infected mice showed no weight loss and behaved the same as the PBS-treated mice (Fig. 4a and b). Taken together, compared to JS/08 virus, JS/09 was more pathogenic for the mammalian host, as evidenced by fatal infection of mice.
Average body weight changes (a) and survival rates (b) of BALB/c mice infected with JS/08 and JS/09 viruses. Six-week-old female BALB/c mice (n=10/group) were inoculated intranasally with 50 μl containing 106 EID50 of the viruses JS/08, JS/09 or PBS (mock), and each group was monitored daily for 14 days. The results are expressed as the mean ± SD
Quantitation of viral RNA loads
Differences in the ability of these viruses to replicate in the organs of mice were also investigated. We chose 3 d.p.i. and 5 d.p.i. to determine the viral RNA loads in different tissues by quantitative PCR. At 3 d.p.i., JS/08 viruses only had detectable viral loads in the lungs of infected mice, with mean titer of 103.3 copies/g. In mice inoculated with the JS/09 strain, the viral RNA titer in the lungs was significantly higher than in JS/08 group, with a mean titer of 107.5 copies/g. The viral RNA loads were also detectable in the heart and spleen of JS/09- infected mice, with a similar mean titer of 103.3 copies/g (Fig. 5a). At 5 d.p.i., no virus was detected in any of the organs of JS/08-infected mice, indicating that the virus might be gradually cleared from the body. However, in the JS/09-infected group, all of the tissues except the kidneys had detectable viral loads. The mean viral RNA titer in the lungs was 108.2 copies/g, which was significantly higher than in the other organs. The heart and spleen still contained similar viral RNA loads, with a mean titer of 103.9 copies/g, whereas the brain and liver contained somewhat lower viral RNA loads, with mean titers of 103.5 copies/g and 103.2 copies/g, respectively (Fig. 5b).
Viral loads in the main organs of mice inoculated with JS/08 and JS/09 viruses at 3 d.p.i. (a) and at 5 d.p.i. (b). Three mice were chosen randomly and euthanized. Viral loads are expressed as log10 RNA copy numbers per gram of sample. The dashed horizontal line indicates the lower limit of detection
Discussion
The Asian HPAI H5N1 viruses emerged over a decade ago in China and then spread into wide range of avian and mammalian species. Here, we reported two cases of swine H5N1 viruses (JS/08 and JS/09) isolated in separate small-scale pig farms in Jiangsu Province of eastern China. Genetic and phylogenetic analysis showed that these two swine H5N1 viruses belonged to clade 7 and clade 2.3.4, respectively. Both of the H5N1 viruses showed a close relationship to the corresponding poultry isolates in the same clade circulating in China during 2007–2009 [13, 18]. Compared to large farms, these small farms practiced relatively poor biosecurity, therefore the H5N1 viruses may be easily transmitted from poultry to pigs in areas with H5N1 virus endemicity and with intermingled raising of poultry and swine. A similar observation was made in a recent surveillance in Indonesia that revealed that 52 swine H5N1 viruses were isolated from pig farms near the HPAI H5N1 outbreak site [25].
However, there was no evidence that JS/08 and JS/09 acquired the ability to transmit from pig to pig, as only one virus was isolated in the same sampling group. In addition, analysis of viral receptor specificities also showed that both of the H5N1 isolates still bound to the avian-type receptor, indicating that they do not adapt when they replicate in the respiratory tract of a pig. The important adaptation of an avian influenza virus to a mammalian host is to switch its preference to bind α2,6-linked SA. To date, most H5 subtype HPAI viruses have retained a binding specificity for α2,3-linked SA and have not demonstrated the capacity for efficient transmission in the ferret and pig models [5, 23, 39]. However, the lack of signs in pigs infected with HPAI H5N1viruses would prolong the time of virus adaption before detection. Moreover, during our surveillance of Jiangsu Province in 2010, eight human A(H1N1)pdm2009 viruses were isolated [41]. A recent study has demonstrated that the reassortant H5 HA/pH1N1 virus could acquire the ability of aerosol transmission in ferrets with a few mutations in the HA gene [11]. Since pigs have long been considered intermediate hosts, the co-circulation of H5N1and A(H1N1)pdm2009 viruses in swine in the same area would increase the likelihood of their reassortment and the subsequent generation of H5N1 viruses that transmit efficiently in humans.
No amino acid substitutions were found at key sites (e.g., E627K and D701N in the PB2 protein) known to be related to virulence in mammalian hosts in either H5N1 isolate [9, 19]. However, compared to JS/08, JS/09 seemed to be of particular concern, as it was obviously more cytopathic in vitro and more pathogenic in mice without prior adaption. These results suggest that the virus belonging to clade 2.3.4 might be transmitted to other mammalian species more easily. Actually, most of the human H5N1 isolates in China belonged to clade 2.3.4, and only one, A/Beijing/01/2003, belonged to clade 7 [35]. The discrepant properties may be explained to some extent by the different multiple basic amino acids at the hemagglutinin cleavage site in these two clades [40]. However, due to the poor surveillance of swine influenza viruses worldwide, the potential risk of pigs becoming a reservoir that could maintain the H5N1 virus or even infect humans is still unclear.
Despite the control efforts that have been made for many years, H5N1 avian influenza is still endemic in China, and the viruses evolve continuously to pose a real threat to the poultry industry and public health [18]. Transmission of H5N1 HPAI viruses from poultry to swine increases the likelihood of human infections and the potential risk of a pandemic. Our findings further demonstrate the important role of pigs as intermediate hosts and their ability be infected naturally with H5N1 viruses, and they emphasize the need to strengthen influenza A virus surveillance in swine.
References
Auewarakul P, Suptawiwat O, Kongchanagul A, Sangma C, Suzuki Y, Ungchusak K, Louisirirotchanakul S, Lerdsamran H, Pooruk P, Thitithanyanont A, Pittayawonganon C, Guo CT, Hiramatsu H, Jampangern W, Chunsutthiwat S, Puthavathana P (2007) An avian influenza H5N1 virus that binds to a human-type receptor. J Virol 81:9950–9955
Bi YH, Fu GH, Chen J, Peng JS, Sun YP, Wang JJ, Pu JA, Zhang Y, Gao HJ, Ma GP, Tian FL, Brown IH, Liu JH (2010) Novel swine influenza virus reassortants in pigs, China. Emerg Inf Dis 16:1162–1164
Castrucci MR, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster RG (1993) Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology 193:503–506
Chen LM, Davis CT, Zhou H, Cox NJ, Donis RO (2008) Genetic compatibility and virulence of reassortants derived from contemporary avian H5N1 and human H3N2 influenza A viruses. PloS Path 4:e1000072
Choi YK, Nguyen TD, Ozaki H, Webby RJ, Puthavathana P, Buranathal C, Chaisingh A, Auewarakul P, Hanh NTH, Ma SK, Hui PY, Guan Y, Peiris JSM, Webster RG (2005) Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J Virol 79:10821–10825
Claas EC, Osterhaus AD, van Beek R, De Jong JC, Rimmelzwaan GF, Senne DA, Krauss S, Shortridge KF, Webster RG (1998) Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472–477
Cong YL, Wang GM, Guan ZH, Chang SA, Zhang QP, Yang GL, Wang WL, Meng QF, Ren WM, Wang CF, Ding ZA (2010) Reassortant between human-like H3N2 and avian H5 subtype influenza A viruses in pigs: a potential public health risk. PloS One 5:e12591
Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ (2009) Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201
Hatta M, Gao P, Halfmann P, Kawaoka Y (2001) Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840–1842
Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR (2001) Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146:2275–2289
Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong GX, Hanson A, Katsura H, Watanabe S, Li CJ, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y (2012) Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428
Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, Kawaoka Y (1998) Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol 72:7367–7373
Jiang WM, Liu S, Chen J, Hou GY, Li JP, Cao YF, Zhuang QY, Li Y, Huang BX, Chen JM (2010) Molecular epidemiological surveys of H5 subtype highly pathogenic avian influenza viruses in poultry in China during 2007–2009. J Gen Virol 91:2491–2496
Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge KF, Kawaoka Y, Webster RG (1994) Potential for transmission of avian influenza viruses to pigs. J Gen Virol 75:2183–2188
Lee MS, Chang PC, Shien JH, Cheng MC, Shieh HK (2001) Identification and subtyping of avian influenza viruses by reverse transcription-PCR. J Virol Methods 97:13–22
Li CJ, Hatta M, Nidom CA, Muramoto Y, Watanabe S, Neumann G, Kawaoka Y (2010) Reassortment between avian H5N1 and human H3N2 influenza viruses creates hybrid viruses with substantial virulence. P Natl Acad Sci USA 107:4687–4692
Li Y, Zhang X, Xu Q, Fu Q, Zhu Y, Chen S, Peng D, Liu X (2013) Characterisation and haemagglutinin gene epitope mapping of a variant strain of H5N1 subtype avian influenza virus. Vet Microbiol 162:614–622
Li YB, Shi JZ, Zhong GX, Deng GH, Tian GB, Ge JY, Zeng XY, Song JS, Zhao DM, Liu LL, Jiang YP, Guan YT, Bu ZG, Chen HL (2010) Continued evolution of H5N1 influenza viruses in wild birds, domestic poultry, and humans in China from 2004 to 2009. J Virol 84:8389–8397
Li Z, Chen H, Jiao P, Deng G, Tian G, Li Y, Hoffmann E, Webster RG, Matsuoka Y, Yu K (2005) Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J Virol 79:12058–12064
Lipatov AS, Kwon YK, Sarmento LV, Lager KM, Spackman E, Suarez DL, Swayne DE (2008) Domestic pigs have low susceptibility to H5N1 highly pathogenic avian influenza viruses. PloS Path 4:e1000102
Liu QF, Ma JJ, Kou Z, Pu J, Lei FM, Li TX, Liu JH (2010) Characterization of a highly pathogenic avian influenza H5N1 clade 2.3.4 virus isolated from a tree sparrow. Virus Res 147:25–29
Liu W, Wei MT, Tong YG, Tang F, Zhang L, Fang LQ, Yang H, Cao WC (2011) Seroprevalence and genetic characteristics of five subtypes of influenza A viruses in the Chinese pig population: a pooled data analysis. Vet J 187:200–206
Maines TR, Chen LM, Matsuoka Y, Chen HL, Rowe T, Ortin J, Falcon A, Hien NT, Mai LQ, Sedyaningsih ER, Harun S, Tumpey TM, Donis RO, Cox NJ, Subbarao K, Katz JM (2006) Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. P Natl Acad Sci USA 103:12121–12126
Negovetich NJ, Webster RG (2010) Thermostability of subpopulations of H2N3 influenza virus isolates from mallard ducks. J Virol 84:9369–9376
Nidom CA, Takano R, Yamada S, Sakai-Tagawa Y, Daulay S, Aswadi D, Suzuki T, Suzuki Y, Shinya K, Iwatsuki-Horimoto K, Muramoto Y, Kawaoka Y (2010) Influenza A (H5N1) viruses from pigs, Indonesia. Emerg Inf Dis 16:1515–1523
Octaviani CP, Ozawa M, Yamada S, Goto H, Kawaoka Y (2010) High level of genetic compatibility between swine-origin H1N1 and highly pathogenic avian H5N1 influenza viruses. J Virol 84:10918–10922
Peiris JSM, de Jong MD, Guan Y (2007) Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev 20:243–267
Qiu BF, Liu WJ, Peng DX, Hu SL, Tang YH, Liu XF (2009) A reverse transcription-PCR for subtyping of the neuraminidase of avian influenza viruses. J Virol Methods 155:193–198
Reed LJ, Muench H (1938) A simple method of estimating fifty percent endpoints. Am J Epidemiol 27:493–497
Shi WF, Gibbs MJ, Zhang YZ, Zhang Z, Zhao XM, Jin X, Zhu CD, Yang MF, Yang NN, Cui YJ, Ji L (2008) Genetic analysis of four porcine avian influenza viruses isolated from Shandong, China. Arch Virol 153:211–217
Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A (2009) Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125
Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N (1998) Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393–396
Suptawiwat O, Kongchanagul A, Chan-It W, Thitithanyanont A, Wiriyarat W, Chaichuen K, Songserm T, Suzuki Y, Puthavathana P, Auewarakul P (2008) A simple screening assay for receptor switching of avian influenza viruses. J Clin Virol 42:186–189
Vijaykrishna D, Smith GJ, Pybus OG, Zhu H, Bhatt S, Poon LL, Riley S, Bahl J, Ma SK, Cheung CL, Perera RA, Chen H, Shortridge KF, Webby RJ, Webster RG, Guan Y, Peiris JS (2011) Long-term evolution and transmission dynamics of swine influenza A virus. Nature 473:519–522
Wan XF, Dong LB, Lan Y, Long LP, Xu CL, Zou SM, Li Z, Wen LY, Cai ZP, Wang W, Li XD, Yuan F, Sui HT, Zhang Y, Dong J, Sun SH, Gao Y, Wang M, Bai T, Yang L, Li DX, Yang WZ, Yu HJ, Wang SW, Feng ZJ, Wang Y, Guo YJ, Webby RJ, Shu YL (2011) Indications that live poultry markets are a major source of human H5N1 influenza virus infection in China. J Virol 85:13432–13438
Xu XY, Subbarao K, Cox NJ, Guo YJ (1999) Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 261:15–19
Yamamoto Y, Nakamura K, Yamada M, Mase M (2012) Limited susceptibility of pigeons experimentally inoculated with H5N1 highly pathogenic avian influenza viruses. J Vet Med Sci 74:205–208
Yee KS, Carpenter TE, Cardona CJ (2009) Epidemiology of H5N1 avian influenza. Comp Immunol Microb 32:325–340
Yen HL, Lipatov AS, Ilyushina NA, Govorkova EA, Franks J, Yilmaz N, Douglas A, Hay A, Krauss S, Rehg JE, Hoffmann E, Webster RG (2007) Inefficient transmission of H5N1 influenza viruses in a ferret contact model. J Virol 81:6890–6898
Zhang Y, Sun YP, Sun HL, Pu J, Bi YH, Shi Y, Lu XS, Li J, Zhu QY, Gao GF, Yang HC, Liu JH (2012) A single amino acid at the hemagglutinin cleavage site contributes to the pathogenicity and neurovirulence of H5N1 influenza virus in mice. J Virol 86:6924–6931
Zhao G, Fan QP, Zhong L, Li YF, Liu WB, Liu XW, Gao S, Peng DX, Liu XF (2012) Isolation and phylogenetic analysis of pandemic H1N1/09 influenza virus from swine in Jiangsu province of China. Res Vet Sci 93:125–132
Zhu Q, Yang H, Chen W, Cao W, Zhong G, Jiao P, Deng G, Yu K, Yang C, Bu Z, Kawaoka Y, Chen H (2008) A naturally occurring deletion in its NS gene contributes to the attenuation of an H5N1 swine influenza virus in chickens. J Virol 82:220–228
Acknowledgments
This study was supported by the Major State Basic Research Development Program (973 Program) (No. 2011CB505003), the Important National Science & Technology Specific Projects (Nos. 2008ZX10004-013 and 2009ZX10004-214), and the Research and Innovation Project of College Students in Jiangsu Province (No.CXZZ12_0914).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
He, L., Zhao, G., Zhong, L. et al. Isolation and characterization of two H5N1 influenza viruses from swine in Jiangsu Province of China. Arch Virol 158, 2531–2541 (2013). https://doi.org/10.1007/s00705-013-1771-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-013-1771-y