Abstract
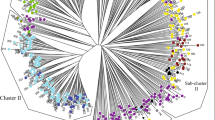
Multiple DNA marker systems and complementary analytical approaches are often useful in population genetic analysis and speciation of plants. We investigated population structure of kenaf (Hibiscus cannabinus) and roselle (H. sabdariffa) for gaining insight in evolution and geographic separation of these crop species using SSR and RGA (resistance gene analogues) markers through Bayesian clustering and principal coordinate analysis (PCoA) methods. Genotyping by 12 SSR and 16 RGA markers amplified a total of 172 loci in the study population. The RGA markers generated higher number of alleles per marker (8.2) as compared to SSR (3.4), but exhibited lower heterozygosity in the population. Genetic variance and heterozygosity in roselle population for both marker systems were lower than in kenaf. RGA markers revealed higher variation among populations. Bayesian structure as well as PCoA analysis using RGA marker revealed distinct cluster for roselle, while SSR-based classification revealed high admixture. Results indicate geographic isolation and natural selection for adaptive RGA alleles in kenaf. The Indian kenaf landraces were distinct from the exotic kenaf accessions, suggesting separate lineage formation by geographic separation. Possible origin and domestication of roselle in South India is proposed.



Similar content being viewed by others
References
Akpan GA, Hossain MG (1998) Karyotypes and evolutionary relations of Hibiscus asper Hook., H. cannabinus L. and H. surattensis L. (Malvaceae). Bot J Linn Soc 126:207–216
Blair MW, Diaz LM, Acosta-Gallegos JA (2013) Race structure in the Mexican collection of common bean landraces. Crop Sci 53:1517–1528
Brugmans B, Wouters D, Van Os H, Hutten R, Linden GVD, Visser RGF, Van Eck HJ, Van Der Vossen EAG (2008) Genetic mapping and transcription analyses of resistance gene loci in potato using NBS profiling. Theor Appl Genet 117:1379–1388
Carvajal-Zarrabal O, Barradas-Dermitz DM, Orta-Flores Z, Hayward-Jones PM, Nolasco-Hipólito C, Aguilar-Uscanga MG, Miranda-Medina A, Bujang KB (2012) Hibiscus sabdariffa L., roselle calyx, from ethnobotany to pharmacology. J Exp Pharmacol 4:25–39
Chen XM, Line RF, Leung H (1998) Genome scanning for resistance-gene analogs in rice, barley, and wheat by high resolution electrophoresis. Theor Appl Genet 97:345–355
Cheng Z, Lu BR, Sameshima K, Fu DX, Chen JK (2004) Identification and genetic relationships of kenaf (Hibiscus cannabinus L.) germplasm revealed by AFLP analysis. Genet Resour Crop Evol 51:393–401
Coetzee R, Herselman L, Labuschagne MT (2008) Genetic diversity analysis of kenaf (Hibiscus cannabinus L.) using AFLP (amplified fragment length polymorphism) markers. Plant Genet Resour Charact Util 7:122–126
Dempsey JM (1964) Kenaf production in Asia. In: Crandall BS (ed) Second international kenaf conference. Department of State Agency for international Development, Washington, D. C., pp 222–238
Dempsey JM (1975) Fibre crops. Rose Printing Company, Tallahassee
Dong P, Wei YM, Chen GY, Li W, Nevo E, Zheng YL (2009) Resistance gene analog polymorphisms (RGAPs) in wild emmer wheat (Triticum dicoccoides) and their ecological associations. Genet Resour Crop Evol 56:121–136
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fuller D, Hildebrand E (2013) Domesticating plants in Africa. In: Mitchell P, Lane P (eds) The Oxford handbook of African archaeology. Oxford University Press, UK, pp 507–526
Garoia F, Guarniero I, Grifoni D, Marzola S, Tinti F (2007) Comparative analysis of AFLPs and SSRs efficiency in resolving population genetic structure of Mediterranean Solea vulgaris. Mol Ecol 16:1377–1387
Gyawali S, Hegedus DD, Parkin IAP, Poon J, Higgins E, Horner K, Bekkaoui D, Coutu C, Buchwaldt L (2013) Genetic diversity and population structure in a world collection of Brassica napus accessions with emphasis on South Korea, Japan, and Pakistan. Crop Sci 53:1537–1545
Hasan M, Seyis F, Badani AG, Pons-Kühnemann J, Friedt W, Lühs W, Snowdon RJ (2006) Analysis of genetic diversity in the Brassica napus L. gene pool using SSR markers. Genet Resour Crop Evol 53:793–802
Jombart T, Pontier D, Dufour AB (2009) Genetic markers in the playground of multivariate analysis. Heredity 102:330–341
Jump AS, Peñuelas J (2007) Extensive spatial genetic structure revealed by AFLP but not SSR molecular markers in the wind-pollinated tree, Fagus sylvatica. Mol Ecol 16:925–936
Kerr HC (1874) Report on the cultivation of, and trade in, jute in Bengal, and on Indian fibres available for the manufacture of paper. Bengal Secretariat Press, Calcutta. http://www.new1.dli.ernet.in/scripts/FullindexDefault.htm?path1=/data3/upload/0078/398&first=1&last=224&barcode=4990010223998
Kuwada H, Mabuchi T (1977) Characteristics of F1 hybrids obtained from the crosses between Hibiscus asper, H. cannabinus and H. sabdariffa (studies on interspecific and intergeneric hybridization in the Malvaceae XIV). Jpn J Breed 27:267–273
Lawrence GJ, Finnegan EJ, Ayliffe MA, Ellis JG (1995) The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco virus resistance gene N. Plant Cell 7:1195–1206
Leister D, Ballvora A, Salamini F, Gebhardt C (1996) A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14:421–429
Liu H, Lin Y, Chen G, Shen Y, Liu J, Zhang S (2012) Genome-scale identification of resistance gene analogs and the development of their intron length polymorphism markers in maize. Mol Breed 29:437–447
Maguire TL, Peakall R, Saenger P (2002) Comparative analysis of genetic diversity in the mangrove species Avicennia marina (Forsk.) Vierh. (Avicenniaceae) detected by AFLPs and SSRs. Theor Appl Genet 104:388–398
Mahapatra BS, Mitra S, Satya P, Satpathy S, Kundu DK, Karmakar PG (2013) Mesta. In: Prasad R (ed) Textbook of fields crops production—commercial crops, vol II. Indian Council of Agricultural Research, New Delhi, pp 399–410
Maiti R (1997) World fibre crops. Science Publisher Inc., Enfield Distribution Co., Enfield
Mandel JR, Dechaine JM, Marek LF, Burke JM (2011) Genetic diversity and population structure in cultivated sunflower and a comparison to its wild progenitor, Helianthus annuus L. Theor Appl Genet 123:693–704
Menzel MY, Martin DW (1971) Chromosome homology in some intercontinental hybrids in Hibiscus sect. Furcaria. Am J Bot 58:191–202
Monti A, Alexopoulou E (2013) Kenaf: a multipurpose crop for several industrial applications, green energy and technology. Springer, London
Muir JP (2002) Effect of dairy compost application and plant maturity on forage kenaf cultivar fiber concentration and in sacco disappearance. Crop Sci 42:248–254
Nishino T, Hiroa AK, Kotera M, Nakamae K, Inagaki H (2003) Kenaf reinforced biodegradable composite. Compos Sci Technol 63:1281–1286
Palomino C, Fernandez-Romero MD, Rubito J, Torres A, Moreno MT (2009) Integration of new CAPS and dCAPS-RGA markers into a composite chickpea genetic map and their association with disease resistance. Theor Appl Genet 118:671–682
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. http://darwin.cirad.fr/darwin. Accessed 12 Oct 2012
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA et al (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909
Pritchard JK, Stevens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falush D (2010) Documentation for structure software: Version 2.3. http://pritch.bsd.uchicago.edu/software/structure_v.2.3.1/documentation.pdf. Accessed 15 Sept 2012
Qi J, Xu J, Li A, Wang X, Zhang G, Su J, Liu A (2011) Analysis of genetic diversity and phylogenetic relationship of kenaf germplasm by SRAP. J Nat Fibers 8:99–110
Rakshit SC, Kundu BC (1970) Revision of the Indian species of Hibiscus. Bull Bot Surv India 12:151–175
Royle JF (1855) The fibrous plants of India fitted for cordage, clothing and paper. Smith, Elder and Co., 65 Cornhill, London
Satya P, Maiti R (2013) Bast and leaf fibre crops: kenaf, hemp, jute, agave etc. In: Singh BP (ed) Biofuel crops: production, physiology and genetics. CAB International, Wallingford, pp 292–311
Satya P, Karan M, Sarkar D, Sinha MK (2012) Genome synteny and evolution of AABB allotetraploids in Hibiscus section Furcaria revealed by interspecific hybridization, ISSR and SSR markers. Plant Syst Evol 298:1257–1270. doi:10.1007/s00606-012-0632-6
Satya P, Karan M, Kar CS, Mahapatra AK, Mahapatra BS (2013) Assessment of molecular diversity and evolutionary relationship of kenaf (Hibiscus cannabinus L.), roselle (H. sabdariffa L.) and their wild relatives. Plant Syst Evol 299:619–629. doi:10.1007/s00606-012-0748-8
Shen J, Araki H, Chen L, Chen JQ, Tian D (2006) Unique evolutionary mechanism in R-genes under the presence/absence polymorphism in Arabidopsis thaliana. Genetics 172:1243–1250
Tuck G, Glendining MJ, Smith P, House JI, Wattenbach M (2006) The potential distribution of bioenergy crops in Europe under present and future climate. Biomass Bioenerg 30:183–197
Varshney RK, Chabane K, Hendre PS, Aggarwal RK, Graner A (2007) Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Sci 173:638–649
Vigouroux Y, Glaubitz JC, Matsuoka Y, Goodman MM, Sánchez JG, Doebley J (2008) Population structure and genetic diversity of new world maize races assessed by DNA microsatellites. Am J Bot 95:1240–1253
Villar JC, Revilla E, Gómez N, Carbajo JM, Simón JL (2009) Improving the use of kenaf for kraft pulping by using mixtures of bast and core fibers. Ind Crop Prod 29:301–307
Wang SP, Liu KD, Wang J, Zhang QF (1998) Identifying candidate disease resistance genes in rice by sequence homology and chromosomal locations. Acta Bot Sin 40:42–50
Webber CL III, Liu A (2011) Plant fibres as renewable feedstocks for biofuel and bio-based products. CCG International, St. Paul
Wilson FD, Menzel MY (1967) Interspecific hybrids between kenaf (Hibiscus cannabinus) and roselle (H. sabdariffa). Euphytica 16:33–44
Woodhead M, Russell J, Squirrell J, Hollingsworth PM, Mackenzie K, Gibby M, Powell W (2005) Comparative analysis of population genetic structure in Athyrium distentifolium (Pteridophyta) using AFLPs and SSRs from anonymous and transcribed gene regions. Mol Ecol 14:1681–1695
Xiao W, Zhao J, Fan S, Li L, Dai J, Xu M (2007) Mapping of genome-wide resistance gene analogs (RGAs) in maize (Zea mays L.). Theor Appl Genet 115:501–508
Yaish MWF, de Miera LES, de la Vega MP (2004) Isolation of a family of resistance gene analogue sequences of the nucleotide binding site (NBS) type from Lens species. Genome 47:650–659
Yu YG, Buss GR, Maroof MAS (1996) Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide-binding site. Proc Natl Acad Sci USA 93:11751–11756
Zhang L, Li A, Wang X, Xu J, Zhang G, Su J, Qi J, Guan C (2013) Genetic diversity of kenaf (Hibiscus cannabinus) evaluated by inter-simple sequence repeat (ISSR). Biochem Genet 51:800–810. doi:10.1007/s10528-013-9608-7
Acknowledgments
We thank Director, Central Research Institute for Jute and Allied Fibres for kind support to the present research work and the reviewers for their constructive comments.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Satya, P., Karan, M., Chakraborty, K. et al. Comparative analysis of diversification and population structure of kenaf (Hibiscus cannabinus L.) and roselle (H. sabdariffa L.) using SSR and RGA (resistance gene analogue) markers. Plant Syst Evol 300, 1209–1218 (2014). https://doi.org/10.1007/s00606-013-0956-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-013-0956-x