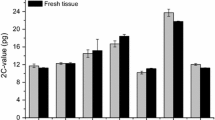
Abstract
Dipsacaceae and Morinaceae have for a long time been regarded as separate but related families, whereas according to APG III they are included within the larger family Caprifoliaceae. Although genome size studies seldom provide conclusive characters for higher level systematics, they can yield useful information at a lower taxonomy level. In this study, we used DNA flow cytometry (supplemented by Feulgen densitometry) for measurement of genome size variation in the Dipsacaceae genera Cephalaria, Dipsacus, Knautia, Lomelosia, Pterocephalus, Scabiosa, Sixalix, Succisa, and Succisella, and Morina of the Morinaceae. At the monoploid level the Dipsacaceae genera (x = 7–10) vary 5.94-fold between 0.902 and 5.362 pg DNA (1Cx-value), whereas Morina longifolia (x = 17) has only 0.670 pg DNA. At the holoploid level 11.58-fold variation occurs between 0.902 and 10.446 pg DNA (1C-value). In Knautia sect. Trichera ploidy levels 2x, 4x, 6x are accompanied by corresponding increments of C-values, but genome downsizing is observed. In Knautia sect. Tricheroides the only investigated species K. integrifolia (2n = 20) has only 0.60-fold the mean genome size of sect. Trichera. Scabiosa canescens (2n = 2x = 16) has approximately double the C-value of all other Austrian Scabiosa species at the diploid level (pseudopolyploidy). These values raise concern against DNA-ploidy estimations at the interspecific level when chromosome numbers are unknown. The species sorted into two major clades of an existing phylogenetic tree of Dipsacaceae differ characteristically in their range of Cx-values. The Knautia–Cephalaria–Dipsacus–Succisella clade has the great majority of its Cx-values larger than those of the Scabiosa–Pterocephalus–Lomelosia clade.


Similar content being viewed by others
References
APG (2003) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc 141:399–436
APG (2009) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc 161:105–121
Avino M, Tortoriello G, Caputo P (2009) A phylogenetic analysis of Dipsacaceae based on four DNA regions. Plant Syst Evol 279:69–86
Backlund A, Bremer B (1997) Phylogeny of the Asteridae s.str. based on rbcL sequences, with particular reference to the Dipsacales. Plant Syst Evol 207:225–254
Bagwell CB, Baker D, Whetstone S, Munson M, Hitchcox S, Ault KA, Lovett EJ (1989) A simple and rapid method for determining the linearity of a flow cytometer amplification system. Cytometry 10:689–694
Baranyi M, Greilhuber J (1996) Flow cytometric and Feulgen densitometric analysis of genome size variation in Pisum. Theor Appl Genet 92:297–307
Bell CD, Edwards EJ, Kim S-T, Donoghue MJ (2001) Dipsacales phylogeny based on chloroplast DNA sequences. Harvard Pap Bot 6:481–499
Benko-Iseppon AM, Morawetz W (2000) Cytological comparison of Calyceraceae and Dipsacaceae with special reference to their taxonomic relationship. Cytologia 65:123–128
Bennett MD (1971) The duration of meiosis. Proc Roy Soc Lond B 178:277–299
Bennett MD (1972) Nuclear DNA content and minimum generation time in herbaceous plants. Proc Roy Soc Lond B 181:109–135
Bennett MD, Leitch IJ (2005a) Angiosperm DNA C-values Database. http://data.kew.org/cvalues
Bennett MD, Leitch IJ (2005b) Plant DNA C-values Database. http://data.kew.org/cvalues
Bennett MD, Leitch IJ (2005c) Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann Bot 95:45–90
Bennetzen JL, Ma J, Devos KM (2005) Mechanisms of recent genome size variation in flowering plants. Ann Bot 95:127–132
Bureš P, Wang Y-F, Horová L, Suda J (2004) Genome size variation in central European species of Cirsium (Compositae) and their natural hybrids. Ann Bot 94:353–363
Caputo P, Cozzolino S (1994) A cladistic analysis of Dipsacaceae (Dipsacales). Plant Syst Evol 189:41–61
Caputo P, Cozzolino S, Moretti A (2004) Molecular phylogenetics of Dipsacaceae reveals parallel trends in seed dispersal syndromes. Plant Syst Evol 246:163–175
Carlson S, Mayer V, Donoghue MJ (2009) Phylogenetic relationships, taxonomy, and morphological evolution in Dipsacaceae (Dipsacales) inferred by DNA sequence data. Taxon 58:1075–1091
Cavalier-Smith T (1985) Eukaryote gene numbers, non-coding DNA and genome size. In: Cavalier-Smith T (ed) The evolution of genome size. Wiley, Chichester, pp 69–103
Doležel J, Greilhuber J, Lucretti S, Meister A, Lysák MA, Nardi L, Obermayer R (1998) Plant genome size estimation by flow cytometry: inter-laboratory comparison. Ann Bot 82(Suppl A):17–26
Donoghue MJ, Eriksson T, Reeves PA, Olmstead RG (2001) Phylogeny and phylogenetic taxonomy of Dipsacales, with special reference to Sinadoxa and Tetradoxa (Adoxaceae). Harvard Pap Bot 6:459–479
Donoghue MJ, Bell CD, Winkworth RC (2003) The evolution of reproductive characters in Dipsacales. Int J Plant Sci 164:S453–S464
Ehrendorfer F (1962) Beiträge zur Phylogenie der Gattung Knautia (Dipsacaceae), I. Cytologische Grundlagen und allgemeine Hinweise. Österr Bot Zeitschr 109:276–343
Ehrendorfer F (1964) Evolution and karyotype differentiation in a family of flowering plants: Dipsacaceae. Genet Today 2:399–407
Feulgen R, Rossenbeck H (1924) Mikroskopisch-chemischer Nachweis einer Nucleinsäure vom Typus der Thymonucleinsäure und die darauf beruhende elektive Färbung von Zellkernen in mikroskopischen Präparaten. Hoppe-Seyler’s Z Physiol Chem 135:203–248
Fischer MA, Oswald K, Adler W (2008) Exkursionsflora für Österreich, Liechtenstein und Südtirol, III edn. Biologiezentrum der Oberösterreichischen Landesmuseen, Linz
Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220:1049–1051
Goldblatt P, Johnson DE (1979-) Index to plant chromosome numbers. http://mobot.mobot.org/W3T/Search/ipcn.html
Gregory TR (2005) The C-value enigma in plants and animals: a review of parallels and an appeal for partnership. Ann Bot 95:133–146
Greilhuber J (1986) Severely distorted Feulgen-DNA amounts in Pinus (Coniferophytina) after nonadditive fixations as a result of meristematic self-tanning with vacuole contents. Can J Genet Cytol 28:409–415
Greilhuber J (1988) “Self-tanning”—a new and important source of stoichiometric error in cytophotometric determination of nuclear DNA content in plants. Plant Syst Evol 158:87–96
Greilhuber J, Ebert I (1994) Genome size variation in Pisum sativum. Genome 37:646–655
Greilhuber J, Temsch EM (2001) Feulgen densitometry: some observations relevant to best practice in quantitative nuclear DNA content determination. Acta Bot Croat 60:285–298
Greilhuber J, Lysák MA, Doležel J, Bennett MD (2005) The origin, evolution and proposed stabilisation of the terms ‘Genome Size’ and ‘C-Value’ to describe nuclear DNA contents. Ann Bot 94:255–260
Greilhuber J, Borsch T, Müller K, Worberg A, Porembski S, Barthlott W (2006) Smallest angiosperm genomes found in Lentibulariaceae, with chromosomes of bacterial size. Plant Biol 8:770–777
Greilhuber J, Temsch EM, Loureiro J (2007) Nuclear DNA content measurement. In: Doležel J, Greilhuber J, Suda J (eds) Flow cytometry with plant cells. Analysis of genes, chromosomes and genomes. Wiley, Weinheim, pp 67–101
Grime JP, Shacklock JML, Band SR (1985) Nuclear DNA contents, shoot phenology and species co-existence in a limestone grassland community. New Phytol 100:435–445
Hofmann U, Göttmann J (1990) Morina L. und Triplostegia Wall. ex DC. im Vergleich mit Valerianaceae und Dipsacaceae. Bot Jahrb Syst 111:499–553
Kachidze N (1929) Karyologische Studien über die Familie der Dipsacaceae. Planta 7:482–502
Kamelina OP (1983) Basic results of the comparative embryological investigation of Dipsacaceae and Morinaceae. In: Erdelská O (ed) Fertilization and embryogenesis in ovulated plants. Veda, Bratislava, pp 343–346
Kamelina OP (1987) Morinaceae and Dipsacaceae. In: Batygina TB, Kamelina OP, Takhtajan AL, Yakovlev MS, Zhukova GY (eds) Comparative embryology of flowering plants. Nauka Publishers, Leningrad, pp 177–192
Kerguélen M (2010) Index Synonymique de la Flore de France. http://www2.dijon.inra.fr/flore-france/ka-kz.htm
Kolář F, Štech M, Trávníček P, Rauchová J, Urfus T, Vít P, Kubešová M, Suda J (2009) Towards resolving the Knautia arvensis agg. (Dipsacaceae) puzzle: primary and secondary contact zones and ploidy segregation at landscape and microgeographic scales. Ann Bot 103:963–974
Leitch IJ, Bennett MD (2004) Genome downsizing in polyploid plants. Biol J Linn Soc 82:651–663
Loureiro J, Rodriguez E, Doležel J, Santos C (2007) Two new nuclear isolation buffers for plant DNA flow cytometry—a test with 37 species. Ann Bot 100:875–888
Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structure reveal recent and rapid genomic DNA loss in rice. Genome Res 14:860–869
Měsíček J (1992) Dipsacaceae. In: Měsíček J, Javůrková-Jarolímová V (eds) List of chromosome numbers of the Czech vascular plants. Academia, Praha, pp 85–87
Otto F, Oldiges H, Göhde W, Jain VK (1981) Flow cytometric measurement of nuclear DNA content variations as a potential in vivo mutagenicity test. Cytometry 2:189–191
Perdetzoglou DK, Kofinas C, Chinou I, Loukis A, Harvala C (2000) A comparative chemotaxonomic study of eight taxa of Dipsacaceae family. Plant Biosyst 134:213–218
Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, Panauld O (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16:1262–1269
Poddubnaja-Arnoldi V (1933) Spermazellen in der Familie der Dipsacaceae. Planta 21:381–386
Suda J, Trávníček P (2006) Reliable DNA ploidy determination in dehydrated tissues of vascular plants by DAPI flow cytometry: new prospects for plant research. Cytometry 69A:273–280
Suda J, Krahulcová A, Trávníček P, Krahulec F (2006) Ploidy level versus DNA ploidy level: an appeal for consistent terminology. Taxon 55:447–450
Temsch EM, Greilhuber J, Krisai R (2010) Genome size in liverworts. Preslia 82:63–80
Verlaque R (1985) Etude biosystématique et phylogénétique des Dipsacaceae. III. Tribus des Knautieae et des Dipsaceae. Rev Cytol Biol Veg Bot 8:171–243
Watson L, Dallwitz MJ (1992 onwards) The families of flowering plants: descriptions, illustrations, identification, and information retrieval. http://delta-intkey.com/angio, Version 25th November 2009
Wendel JF, Cronn RC, Johnston JS, Price HJ (2002) Feast and famine in plant genomes. Genetics 115:37–47
Yokoya K, Roberts AV, Mottley J, Lewis R, Brandham PE (2000) Nuclear DNA amounts in Roses. Ann Bot 85:557–561
Zhang W-H, Chen Z-D, Li J-H, Chen H-B, Tang Y-C (2003) Phylogeny of the Dipsacales s.l. based on chloroplast trnL-F and ndhF sequences. Mol Phylogen Evol 26:176–189
Acknowledgments
The authors thank Friedrich Ehrendorfer, Luise Schratt-Ehrendorfer, Elvira Hörandl, Veronika Mayer, Peter Schönswetter, Birgit Taumberger, and Karin Tremetsberger for providing material, and Dessislava Dimitrova, Ana Petrova, and Vladimir Vladimirov for helping with the Bulgarian species collection and identification. Thanks also to J. Doležel for standard seeds of S. cereale, H. vulgare and V. faba.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Temsch, E.M., Greilhuber, J. Genome size in Dipsacaceae and Morina longifolia (Morinaceae). Plant Syst Evol 289, 45–56 (2010). https://doi.org/10.1007/s00606-010-0330-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-010-0330-1