Abstract
The authors describe a low-cost and sensitive method for the fluorometric determination of Salmonella enteritidis (S. enteritidis). It is based on the use of nanographite (NG) as a quencher of fluorescence, and on cyclic signal amplification by using deoxyribonuclease I (DNase I). The probe containing a capture probe and two short sequences (probe 1 and probe 2) labeled with carboxyfluorescein (FAM) are used as the signal probe that is adsorbed on the surface of NG via p-stacking interactions. Adsorption results in quenching of the fluorescence of FAM. If S. enteridis is introduced, fluorescence is restored due to the displacement of the probe from the surface of the NG due competitive binding. The signal is considerably amplified by applying DNase I-mediated target recycling. The assay has a linear response that covers the 1 to 50 nM concentration range, with a 0.5 nM lower limit of detection (LOD). A milk sample spiked with S. enteritidis was analyzed and the detection limit is as low as 50 colony-forming units (CFU) per mL. Accordingly, this biosensor is highly sensitive and selective but also cost-effective. Conceivably, the detection scheme may be extended to the detection of other biomolecules.

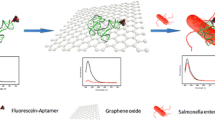
Schematic of the fluorescent strategy for Salmonella enteritidis assay by using nanographite as a quencher and deoxyribonuclease I-aided signal amplification





Similar content being viewed by others
References
Oliver SP, Jayarao BM, Almeida RA (2005) Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodborne Pathog Dis 2:115–129
Nygård K, Jong BD, Guerin PJ, Andersson Y, Olsson A, Giesecke J (2004) Emergence of new Salmonella enteritidis phage types in Europe? Surveillance of infections in returning travelers. BMC Med 2:1–8
Forshell LP , Wierup M (2006) Salmonella contamination: a significant challenge to the global marketing of animal food products. Rev Sci Tech 25:541–554
Pui CF, Wong WC, Chai LC, Tunung R, Jeyaletchumi P, Hidayah MS, Ubong A, Farinazleen MG, Cheah YK, Son R (2011) Salmonella: a foodborne pathogen. Int Food Res J 18:465–473
Gómez-Aldapa CA, Rangel-Vargas E, León HB, Filardo-Kerstupp RS, Gordillo-Martínez AJ, Vázquez-Barrios ME (2013) Presence of diarrheagenic Escherichia coli pathotypes, salmonella and indicator bacteria on fresh-cut jicama [Pachyrhizus erosus (l.) urban] from public markets in mexico. Afr J Microbiol Res 7:4323–4330
Bakthavathsalam P, Rajendran VK, Saran U, Chatterjee S, Ali BM (2013) Immunomagnetic nanoparticle based quantitative PCR for rapid detection of salmonella. Microchim Acta 180:1241–1248
Lee SH, Jung BY, Rayamahji N, Lee HS, Jeon WJ, Choi KS, Kweon CH, Yoo HS (2009) A multiplex real-time PCR for differential detection and quantification of Salmonella spp., Salmonella enterica serovar Typhimurium and Enteritidis in meats. J Vet Sci 10:43–51
Paião FG, Arisitides LG, Murate LS, Vilasbôas GT, Vilasboas LA, Shimokomaki M (2013) Detection of salmonella spp, salmonella enteritidis and typhimurium in naturally infected broiler chickens by a multiplex PCR-based assay. Braz J Microbiol 44:37–41
Priyanka B, Patil RK, Dwarakanath S (2016) A review on detection methods used for foodborne pathogens. Indian J Med Res 144:327–338
Vidic J, Manzano M, Chang CM, Jaffrezic-Renault N (2017) Advanced biosensors for detection of pathogens related to livestock and poultry. Vet Res 48:11
Bang J, Shukla S, Kim YH, Kim M (2012) Development of indirect competitive elisa for the detection of salmonella typhimurium. Rom Biotechnol Lett 17:7194–7204
Chunglok W, Wuragil DK, Oaew S, Somasundrum M, Surareungchai W (2011) Immunoassay based on carbon nanotubes-enhanced elisa for salmonella enterica, serovar typhimurium. Biosens Bioelectron 26:3584–3589
Fei J, Dou W, Zhao G (2016) Amperometric immunoassay for the detection of salmonella pullorum, using a screen-printed carbon electrode modified with gold nanoparticle-coated reduced graphene oxide and immunomagnetic beads. Microchim Acta 183:1–8
Fei J, Dou W, Zhao G (2015) A sandwich electrochemical immunosensor for salmonella pullorum and salmonella gallinarum based on a screen-printed carbon electrode modified with an ionic liquid and electrodeposited gold nanoparticles. Microchim Acta 182:2267–2275
Zhang Q, Chen T, Yang S, Wang X, Guo H (2013) Response surface methodology to design a selective enrichment broth for rapid detection of salmonella spp. by sybr green Ι real-time PCR. Appl Microbiol Biotechnol 97:4149–4158
Liu K, Yan X, Mao B, Wang S, Deng L (2016) Aptamer-based detection of salmonella enteritidis, using double signal amplification by klenow fragment and dual fluorescence. Microchim Acta 183:643–649
Yang S, Li W, Duan Y, Li Z, Le D (2014) Nicking enzyme-assisted biosensor for salmonella enteritidis detection based on fluorescence resonance energy transfer. Biosens Bioelectron 55:400–404
Ning Y, Li WK, Duan YF, Yang M, Deng L (2014) High specific DNAzyme-aptamer sensor for Salmonella paratyphi A using single-walled nanotubes-based dual fluorescence-spectrophotometric methods. J Biomol Screen 19:1099–1106
Sasaki H, Kawamoto E, Okiyama E, Ueshiba H, Mikazuki K, Amao H, Sawada T (2006) Molecular typing of pasteurella pneumotropica isolated from rodents by amplified 16S ribosomal DNA restriction analysis and pulsed-field gel electrophoresis. Microbiol Immunol 50:265–272
Mériaux A, Harvey I, Cormier Y, Beaulieu C, Akimov VN, Duchaine C (2001) Random amplified ribosomal DNA restriction analysis for rapid identification of thermophilic actinomycete-like bacteria involved in hypersensitivity pneumonitis. Syst Appl Microbiol 24:277–284
Piao Y, Liu F, Seo TS (2012) A novel molecular beacon bearing a graphite nanoparticle as a nanoquencher for in situ mRNA detection in cancer cells. ACS Appl Mater Interfaces 4:6785–6789
Wei Y, Li B, Wang X, Duan Y (2014) A nano-graphite-DNA hybrid sensor for magnified fluorescent detection of mercury(ii) ions in aqueous solution. Analyst 139:1618–1621
Pérez-López B, Merkoçi A (2012) Carbon nanotubes and graphene in analytical sciences. Microchim Acta 179:1–16
Ning Y, Li ZJ, Duan YF, Peng ZH, Deng L (2013) A novel biosensor for detection of salmonella typhimurium carrying SSeC gene based on the secondary quenching effect of carbon nanotubes. J Nanomater Mol Nanotechnol 2:1–5
Duan YF, Ning Y, Song Y, Deng L (2014) Fluorescent aptasensor for the determination of salmonella typhimurium, based on a graphene oxide platform. Microchim Acta 181:647–653
Lu Z, Chen X, Wang Y, Zheng X, Li CM (2015) Aptamer based fluorescence recovery assay for aflatoxin b1 using a quencher system composed of quantum dots and graphene oxide. Microchim Acta 182:571–578
Wei Y, Zhang J, Wang X1, Duan Y (2015) Amplified fluorescent aptasensor through catalytic recycling for highly sensitive detection of ochratoxin a. Biosens Bioelectron 65: 16–22
Wei Y, Li B, Wang X, Duan Y (2014) Magnified fluorescence detection of silver(I) ion in aqueous solutions by using nano-graphite-DNA hybrid and DNase I. Biosens Bioelectron 58:276–281
Huang Y, Chen J, Zhao S, Shi M, Chen ZF, Liang H (2013) Label-free colorimetric aptasensor based on nicking enzyme assisted signal amplification and dnazyme amplification for highly sensitive detection of protein. Anal Chem 85:4423–4430
Bao T, Shu H, Wei W, Zhang X, Wang S (2014) A sensitive electrochemical aptasensor for atp detection based on exonuclease III-assisted signal amplification strategy. Anal Chim Acta 862:64–69
Xuan F, Luo X, Hsing I (2012) Ultrasensitive solution-phase electrochemical molecular beacon-based DNA detection with signal amplification by exonuclease III-assisted target recycling. Anal Chem 84:5216–5220
Niazov T, Pavlov V, Xiao Y, Gill R, Willner I (2004) DNAzyme-functionalized AU nanoparticles for the amplified detection of DNA or telomerase activity. Nano Lett 4:1683–1687
Yi H, Xu W, Yuan Y, Bai L, Wu Y, Chai Y, Yuan R (2014) A pseudo triple-enzyme cascade amplified aptasensor for thrombin detection based on hemin/g-quadruplex as signal label. Biosens Bioelectron 54:415–420
Xie Y, Lin X, Huang Y, Pan R, Zhu Z, Zhou L, Yang CJ (2015) Highly sensitive and selective detection of miRNA: DNase I-assisted target recycling using DNA probes protected by polydopamine nanospheres. Chem Commun 51:2156–2158
Lei P, Tang H, Ding S, Ding X, Zhu D, Shen B, Shen B, Cheng Q, Yan Y (2015) Determination of the invA, gene of salmonella, using surface plasmon resonance along with streptavidin aptamer amplification. Microchim Acta 182:289–296
Luo R, Li Y, Lin X, Dong F, Zhang W, Yan L, Cheng W, Ju H, Ding S (2014) A colorimetric assay method for invA, gene of salmonella, using DNAzyme probe self-assembled gold nanoparticles as single tag. Sensors Actuators B Chem 198:87–93
Chen YY, Fang YC, Lin SY, Lin YJ, Yen SY, Huang CH, Yang CY, Chau LK, Wang SC (2017) Corona-induced micro-centrifugal flows for concentration of Neisseria and Salmonella bacteria prior to their quantitation using antibody-functionalized SERS-reporter nanobeads. Microchim Acta 184:1021–1028
Acknowledgments
We would like to thank National Natural Science Foundation of Hunan province(2016JJ3098), Outstanding Youth Scientific Research Project Funded by Education department of Hunan Province (15B169), National Natural Science Foundation (81202451), Science and Technology Innovation Team in Colleges and Universities in Hunan Province《Chinese traditional medicine for treatment of infectious diseases》(No: 15, Grxjb-7), Doctor start-up Foundation of Hunan University of Chinese Medicine (9982-1001019), Key Subjects of Hunan University of Chinese Medicine《pathogenic biology》(NO.1) and Project Funded by Hunan Provincial Level Course《Immunology and pathogenic biology》(No. 48) for the financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author(s) declare that they have no competing interests.
Electronic supplementary material
ESM 1
(PDF 258 kb)
Rights and permissions
About this article
Cite this article
He, Q., Luo, H., Tang, L. et al. Nanographite-based fluorescent biosensing of Salmonella enteritidis by applying deoxyribonuclease-assisted recycling. Microchim Acta 184, 3875–3882 (2017). https://doi.org/10.1007/s00604-017-2363-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-017-2363-4