Abstract
The use of microcentrifugal flows driven by ionic wind that is generated near a corona needle tip above the level of a small liquid reservoir is a technique to trap suspended bacteria within a few minutes. Gas ions are ejected at the tip of a corona needle to form an ionic wind that can swipe across the liquid/air interface inside a miniaturized reservoir to generate centrifugal vortices. This will drag suspended particles to the center of the reservoir. This article describes a method where antigen-functionalized polystyrene microspheres are first employed to mimic bacteria. The mimetic bacteria were then detected by surface-enhanced Raman scattering (SERS) signal after binding antibody-conjugated nanoaggregate-embedded beads (NAEBs) to antigen-functionalized polystyrene microspheres. The NAEBs were prepared from silica-coated gold nanoparticle aggregates and labeled with Raman reporter molecules. The work demonstrates that the particles can be concentrated at the stagnant point at the bottom of a reservoir containing 60 μL solution only by ionic wind-driven centrifugal flows. SERS signals were acquired to identify the bacteria-mimicking particles. The ionic wind flows were then applied to trap and concentrate Neisseria and Salmonella bacteria bound with antibody-conjugated NAEBs at the level of 106 colony forming units (CFU) per mL (or lower) within 10 min. The NAEBs were preferably labeled with Ethyl violet or fluorescein derivatives. The log-log calibration plot for Neisseria is linear in the 104 to 106 CFU⋅mL−1 concentration range.

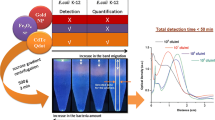
Bacteria bound with antibody-functionalized SERS micro-tags are concentrated with corona-induced micro-centrifugal flows. Trapped bacteria at the reservoir bottom are recognized and quantified using Raman microscope based on SERS signals from the tags.




Similar content being viewed by others
References
Dineva MA, Mahilum-Tapay L, Lee H (2007) Sample preparation: a challenge in the development of point-of-care nucleic acid-based assays for resource-limited settings. Analyst 132(12):1193–1199
Pal N, Sharma S, Gupta S (2016) Sensitive and rapid detection of pathogenic bacteria in small volumes using impedance spectroscopy technique. Biosens Bioelectron 77:270–227
Thiramanas R, Laocharoensuk R (2016) Competitive binding of polyethyleneimine-coated gold nanoparticles to enzymes and bacteria: a key mechanism for low-level colorimetric detection of gram-positive and gram-negative bacteria. Microchim Acta 183(1):389–396
Duan N, Wu S, Dai S, Miao T, Chen J, Wang Z (2015) Simultaneous detection of pathogenic bacteria using an aptamer based biosensor and dual fluorescence resonance energy transfer from quantum dots to carbon nanoparticles. Microchim Acta 182(5–6):917–923
Wang B, Wang Q, Cai Z, Ma M (2015) Simultaneous, rapid and sensitive detection of three food-borne pathogenic bacteria using multicolor quantum dot probes based on multiplex fluoroimmunoassay in food samples. LWT-Food Sci Technol 61(2):368–376
Dogan Ü, Kasap E, Cetin D, Suludere Z, Boyaci IH, Türkyılmaz C, Ertas N, Tamer U (2016) Rapid detection of bacteria based on homogenous immunoassay using chitosan modified quantum dots. Sensors Actuators B Chem 233:369–378
Wu J, Ben Y, Battigelli D, Chang H-C (2005) Long-range AC electroosmotic trapping and detection of Bioparticles. Ind Eng Chem Res 44(8):2815–2822
Lui C, Cady N, Batt C (2009) Nucleic acid-based detection of bacterial pathogens using integrated microfluidic platform systems. Sensors 9(5):3713
Voldman J (2006) Electrical forces for microscale cell manipulation. Annu Rev Biomed Eng 8(1):425–454
Cheng I-F, Chen T-Y, Lu R-J, Wu H-W (2014) Rapid identification of bacteria utilizing amplified dielectrophoretic force-assisted nanoparticle-induced surface-enhanced Raman spectroscopy. Nanoscale Res Lett 9(1):1–8
Chrimes AF, Khoshmanesh K, Tang S-Y, Wood BR, Stoddart PR, Collins SSE, Mitchell A, Kalantar-zadeh K (2013) In situ SERS probing of Nano-silver coated individual yeast cells. Biosens Bioelectron 49:536–541
Cheng IF, Chang H-C, Chen T-Y, Hu C, Yang F-L (2013) Rapid (<5 min) identification of pathogen in human blood by Electrokinetic concentration and surface-enhanced Raman spectroscopy. Sci Report 3:2365
Pohl HA (1978) Dielectrophoresis. Cambridge University Press, London
Kohlheyer D, Besselink GAJ, Schlautmann S, Schasfoort RBM (2006) Free-flow zone electrophoresis and isoelectric focusing using a Microfabricated glass device with ion permeable membranes. Lab Chip 6(3):374–380
Lu H, Gaudet S, Schmidt MA, Jensen KF (2004) A Microfabricated device for subcellular organelle sorting. Anal Chem 76(19):5705–5712
Song Y-A, Chan M, Celio C, Tannenbaum SR, Wishnok JS, Han J (2010) Free-flow zone electrophoresis of peptides and proteins in PDMS microchip for narrow pI range sample Prefractionation coupled with mass spectrometry. Anal Chem 82(6):2317–2325
Kohler S, Weilbeer C, Howitz S, Becker H, Beushausen V, Belder D (2011) PDMS free-flow electrophoresis chips with integrated partitioning bars for bubble segregation. Lab Chip 11(2):309–314
Yeo LY, Hou D, Maheshswari S, Chang H-C (2006) Electrohydrodynamic surface Microvortices for mixing and particle trapping. Appl Phys Lett 88(23):233512
Hou D, Maheshwari S, Chang H-C (2007) Rapid Bioparticle concentration and detection by combining a discharge driven vortex with surface enhanced Raman scattering. Biomicrofluidics 1(1):014106
Smith E, Dent G (2013) Modern Raman spectroscopy: a practical approach. John Wiley & Sons, Chichester
Li D-W, Zhai W-L, Li Y-T, Long Y-T (2014) Recent progress in surface enhanced Raman spectroscopy for the detection of environmental pollutants. Microchim Acta 181(1–2):23–43
Krafft C, Popp J (2015) The many facets of Raman spectroscopy for biomedical analysis. Anal Bioanal Chem 407(3):699–717
Syed MA, Bokhari S (2011) Gold nanoparticle based microbial detection and identification. J Biomed Nanotechnol 7(2):229–237
Pahlow S, Meisel S, Cialla-May D, Weber K, Rösch P, Popp J (2015) Isolation and identification of bacteria by means of Raman spectroscopy. Adv Drug Deliv Rev 89:105–120
Zhou H, Yang D, Mircescu NE, Ivleva NP, Schwarzmeier K, Wieser A, Schubert S, Niessner R, Haisch C (2015) Surface-enhanced Raman scattering detection of bacteria on microarrays at single cell levels using silver nanoparticles. Microchim Acta 182(13–14):2259–2266
Huang P-J, Tay L-L, Tanha J, Ryan S, Chau L-K (2009) Single-domain antibody-conjugated nanoaggregate-embedded beads for targeted detection of pathogenic bacteria. Chem Eur J 15(37):9330–9334
Lin H-Y, Huang C-H, Hsieh W-H, Liu L-H, Lin Y-C, Chu C-C, Wang S-T, Kuo IT, Chau L-K, Yang C-Y (2014) On-line SERS detection of single bacterium using novel SERS nanoprobes and a microfluidic dielectrophoresis device. Small 10(22):4414–4414
Huang P-J, Chau L-K, Yang T-S, Tay L-L, Lin T-T (2009) Nanoaggregate-embedded beads as novel Raman labels for Biodetection. Adv Funct Mater 19(2):242–248
Tay L-L, Huang P-J, Tanha J, Ryan S, Wu X, Hulse J, Chau L-K (2012) Silica encapsulated SERS Nanoprobe conjugated to the bacteriophage Tailspike protein for targeted detection of salmonella. Chem Commun 48(7):1024–1026
Wu X, Han C, Chen J, Huang Y-W, Zhao Y (2016) Rapid detection of pathogenic bacteria from fresh produce by filtration and surface-enhanced Raman spectroscopy. JOM 68(4):1156–1162
Madiyar FR, Bhana S, Swisher LZ, Culbertson CT, Huang X, Li J (2015) Integration of a nanostructured dielectrophoretic device and a surface-enhanced Raman probe for highly sensitive rapid bacteria detection. Nanoscale 7(8):3726–3736
Acknowledgements
The authors thank the grant support from the Ministry of Science and Technology, Taiwan.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The author(s) declare that they have no competing interests.
Electronic supplementary material
ESM 1
(DOC 38 kb)
Rights and permissions
About this article
Cite this article
Chen, YY., Fang, YC., Lin, SY. et al. Corona-induced micro-centrifugal flows for concentration of Neisseria and Salmonella bacteria prior to their quantitation using antibody-functionalized SERS-reporter nanobeads. Microchim Acta 184, 1021–1028 (2017). https://doi.org/10.1007/s00604-017-2077-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-017-2077-7