Abstract
Skin cancer affects the lives of millions of people every year, as it is considered the most popular form of cancer. In the USA alone, approximately three and a half million people are diagnosed with skin cancer annually. The survival rate diminishes steeply as the skin cancer progresses. Despite this, it is an expensive and difficult procedure to discover this cancer type in the early stages. In this study, a threshold-based automatic approach for skin cancer detection, classification, and segmentation utilizing a meta-heuristic optimizer named sparrow search algorithm (SpaSA) is proposed. Five U-Net models (i.e., U-Net, U-Net++, Attention U-Net, V-net, and Swin U-Net) with different configurations are utilized to perform the segmentation process. Besides this, the meta-heuristic SpaSA optimizer is used to perform the optimization of the hyperparameters using eight pre-trained CNN models (i.e., VGG16, VGG19, MobileNet, MobileNetV2, MobileNetV3Large, MobileNetV3Small, NASNetMobile, and NASNetLarge). The dataset is gathered from five public sources in which two types of datasets are generated (i.e., 2-classes and 10-classes). For the segmentation, concerning the “skin cancer segmentation and classification” dataset, the best reported scores by U-Net++ with DenseNet201 as a backbone architecture are 0.104, \(94.16\%\), \(91.39\%\), \(99.03\%\), \(96.08\%\), \(96.41\%\), \(77.19\%\), \(75.47\%\) in terms of loss, accuracy, F1-score, AUC, IoU, dice, hinge, and squared hinge, respectively, while for the “PH2” dataset, the best reported scores by the Attention U-Net with DenseNet201 as backbone architecture are 0.137, \(94.75\%\), \(92.65\%\), \(92.56\%\), \(92.74\%\), \(96.20\%\), \(86.30\%\), \(92.65\%\), \(69.28\%\), and \(68.04\%\) in terms of loss, accuracy, F1-score, precision, sensitivity, specificity, IoU, dice, hinge, and squared hinge, respectively. For the “ISIC 2019 and 2020 Melanoma” dataset, the best reported overall accuracy from the applied CNN experiments is \(98.27\%\) by the MobileNet pre-trained model. Similarly, for the “Melanoma Classification (HAM10K)” dataset, the best reported overall accuracy from the applied CNN experiments is \(98.83\%\) by the MobileNet pre-trained model. For the “skin diseases image” dataset, the best reported overall accuracy from the applied CNN experiments is \(85.87\%\) by the MobileNetV2 pre-trained model. After computing the results, the suggested approach is compared with 13 related studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Skin cancer is the abnormal growth of cells located in the skin that frequently develop on the sun-exposed (i.e., UV light) regions of the skin. However, skin regions that are not regularly exposed to sunlight may develop cancer. Around the world, skin cancer is the most prevalent cancer type. The primary types of skin cancer include “basal cell carcinoma,” “squamous cell carcinoma,” and “melanoma.” Every year, the number of diagnosed cases is over 3.5 million in the United States, which exceeds the counts of lung, breast, and colon cancers combined. Every 57 seconds, one person is diagnosed with skin cancer [48].
Skin cancer can be categorized into two major categories (i.e., melanoma and non-melanoma) concerning the cell type that developed cancer. The different types of each category, statistics, risk factors, diagnosis, and treatment will be discussed in Sect. 2.1. Early detection and screening of skin cancer can lead to a full recovery, as it is with every cancer type. Different machine and deep learning architectures and approaches had been proposed to perform the task of skin cancer detection, classification, and segmentation, e.g., support vector machines SVM [24], fuzzy C-means [116], recurrent neural networks [111], and deep neural networks [35].
The current study concentrates on skin cancer detection, classification, and segmentation. The classification is accomplished using 8 pre-trained convolution neural network (CNN) models. They are VGG16, VGG19, MobileNet, MobileNetV2, MobileNetV3Large, MobileNetV3Small, NASNetMobile, and NASNetLarge. The segmentation is done by employing five different U-Net models. They are U-Net, U-Net++, Attention U-Net, V-Net, and Swin U-Net. Additionally, the CNN hyperparameters optimization is done using the sparrow search algorithm (SpaSA) to gain the performance metrics of the state-of-the-art (SOTA).
1.1 Paper contributions
The current study contributions can be recapped in the next points:
-
Presenting a survey on the skin disorders with a graphical taxonomy.
-
Performing the skin cancer segmentation task using U-Net, U-Net++, Attention U-Net, V-Net, and swin U-Net models.
-
Performing the skin cancer classification task using 8 pre-trained CNN models (i.e., VGG16, VGG19, MobileNet, MobileNetV2, MobileNetV3Large, MobileNetV3Small, NASNetMobile, NASNetLarge).
-
Utilizing the SpaSA approach for the hyperparameters optimization processes.
-
Reporting the SOTA performance metrics and comparing them with different related studies and approaches.
1.2 Paper organization
The rest of the current study is organized as follows: Section 2 presents a survey of skin disorders. Section 3 presents and summarizes the related studies. In Section 4, the background is discussed. It represents deep learning classification, parameters optimization, transfer learning, data scaling, data augmentation, segmentation, deep learning segmentation, meta-heuristic optimization, and performance metrics. In Section 5, a discussion about the methodology, datasets acquisition and pre-processing segmentation phase, learning and optimization, and the overall pseudo-code is discussed. Section 6 presents the details and discussions of the experiments and results. Section 7 presents the study limitations, and finally, Section 8 concludes the paper and presents the future work.
2 Skin disorders medical survey
The largest organ in the body is the skin [133]. It helps regulate body temperature, protects against injuries (and infections), produces vitamin D, and stores fat (and water). It consists of three main layers. They are (1) Epidermis (i.e., the skin outer layer), (2) Dermis (i.e., the skin inner layer), and (3) Hypo-dermis (i.e., the skin deepest layer). Skin disorders are the conditions that affect the layers of the skin [57]. They can cause rashes, sores, itching, or other changes. Some skin conditions can be caused by lifestyle factors, while others can be a result of genetic factors. Skin disorders differ widely in severity and symptoms, as they can be genetic or situational causes; permanent or temporary; painful or painless; and life-threatening or minor [3].
2.1 Skin disorders taxonomy
Skin-related disorders can be classified into permanent and temporary. Temporary disorders include (1) acne, (2) contact dermatitis, (3) cold sore, (4) keratosis pilaris, (5) blister, (6) hives, (7) sunburn, (8) actinic keratosis, (9) carbuncle, (10) latex allergy, (11) cellulitis, (12) measles, (13) chickenpox, and (14) impetigo. Permanent disorders can be divided into skin cancer and skin diseases (i.e., not cancer). Skin diseases are (1) lupus, (2) eczema, (3) rosacea, (4) seborrheic dermatitis, (5) psoriasis, (6) vitiligo, and (7) melasma. As mentioned before, melanoma and non-melanoma are the two main categories of skin cancer [101]. Types of melanoma cancers incorporate (1) superficial spreading melanoma, (2) nodular melanoma, (3) acral lentiginous melanoma, (4) lentigo maligna melanoma, and (5) other rare melanomas. Squamous cell carcinoma, basal cell carcinoma, Merkel cell cancer, and cutaneous lymphomas are types of non-melanoma skin cancer [8, 45]. Figure 1 shows a taxonomy of skin disorders with graphical samples.
2.2 Melanoma skin cancer
Melanoma is one of the deadly cancers in the world. It may spread to other body parts if it has not been discovered and treated in an early phase. In the following subsections, the melanoma development, statistics, and other information related to it will be presented.
2.2.1 Melanoma development
The most profound epidermis layer found exactly above the dermis has the melanocytes cells which produce the color (i.e., pigment) of the skin. When healthy melanocytes get out of control, a cancerous tumor (i.e., melanoma) is created [25, 102]. This tumor can extend to further body parts. Occasionally, normal moles or nevi already located on the skin can form a melanoma. In such a case, the mole undergoes changes that are usually visible (e.g., shape, border, size, or the color of the mole changing) [92]. Scalp, face, trunk, or torso (i.e., abdomen, back, and chest), arms, and legs are the most common melanoma locations. Nonetheless, it can develop on the neck, head, and areas that are not exposed to the sun (i.e., anywhere on the body).
Cutaneous melanoma is considered as the most common type of melanoma type that develops first in the skin. There are three popular types of it. First, “superficial spreading melanoma” is the most popular one that presents up to \(70\%\) of melanomas and commonly develops from a present mole. The second one is “lentigo maligna melanoma” that older people are more likely to develop. Frequently, it begins on skin areas that are often sun-exposed. About \(15\%\) of melanomas are diagnosed as “nodular melanoma” representing the third melanoma type [83]. It usually arises as a bump on the skin.
More rarely, the mouth, the mucous membranes that line the gastrointestinal tract, and a woman’s vagina can develop melanoma. Also, melanoma can develop in the eye [26]. It is worth mentioning that the rare types are reported graphically in Fig. 1.
2.2.2 Melanoma Statistics
In the USA, an estimated number of 106, 110 adults (i.e., 62, 260 men and 43, 850 women) are diagnosed with invasive melanoma in 2021 [29, 114, 115]. Among men and women, the fifth most common type of cancer is melanoma. It is less common in black people than in white people by 20 times and is one of the most regular types of cancer diagnosed among young adults, specifically women. The average diagnostic age for it is 65. In 2020, the predicted melanoma cases diagnosed in people aged 15 to 29 were about 2, 400. Over the past three decades, the number of diagnosed cases with this type of cancer has increased sharply [44].
The rates increased annually by around \(2\%\) from 2008 to 2017. However, the number of melanoma-diagnosed teenagers aged 15 to 19 between 2007 and 2016 declined by \(6\%\) a year. The number of adults in their 20’s is decreased by \(3\%\). Most of the deaths associated with skin cancer are caused by melanoma and approximately represent \(75\%\) of deaths. 7, 180 deaths (i.e., 4, 600 men and 2, 580 women) are estimated to occur in 2021 from melanoma. However, deaths from melanoma have decreased from 2014 to 2018 by almost \(5\%\) in adults older than 50 and by \(7\%\) otherwise [114].
2.2.3 Melanoma risk factors
Risk factors for melanoma include indoor tanning (i.e., the people using sun lamps, tanning beds, or tanning parlors are more likely to grow all types of skin cancer) [95], moles, and fair skin (i.e., people with blue eyes, blond or red hair, and freckles). Studies show that \(10\%\) of melanomas may be linked to genetic factors or conditions [74]. Some inherited genetic conditions, previous skin cancer, race or ethnicity, weakened or suppressed immune system are also risk factors for melanoma [84].
2.2.4 Melanoma early recognition
Recognition of the early warning signs [84] including new skin growth, a suspicious change in an existing mole or nevi, and a non-healing sore in two weeks is very important. Changes in the mole size, shape, feel, or color are often the initial and most important warning signs of melanoma [41, 84, 92]. A guide to the familiar signs of melanoma is known as the “ABCDE” rule [125] and can be summarized as follows:
-
A is for asymmetry: One half of a nevus or mole does not match the other.
-
B is for border: The edges are blurred, notched, ragged, or irregular.
-
C is for color: The color of the mole varies and may have black, brown, and tan shades and white, gray, red, or blue areas.
-
D is for diameter: The spot is larger than 6 millimeters across (i.e., about 0.25 inch), although melanomas sometimes can be more diminutive than this.
-
E is for evolving: The size, color, or shape of a mole is altered. Additionally, when an existing nevus develops melanoma, its texture becomes hard or lumpy.
2.2.5 Melanoma diagnosis and treatment
For melanoma, a biopsy from the lesion (i.e., the suspicious area of skin) is the only sure way to diagnose the cancer [43]. During a biopsy, a sample of tissue is possessed to be tested in a laboratory. Computed tomography scan, ultrasound, positron emission tomography scan, and magnetic resonance imaging are some of the other tests that can be done to diagnose and define the stage of melanoma [47].
Treatment recommendations are depending on numerous factors, counting the stage of the melanoma, the thickness of the initial melanoma, whether cancer has grown or not, rate of melanoma growth, the presence of specified genetic changes in the affected cells, and some other medical circumstances [84]. To obtain a suitable treatment arrangement, potential side effects, the overall health, and preferences of the patient are considered [122].
For people with local melanoma and most people with regional one, the main treatment is surgery. Radiation therapy is another treatment option that employs X-rays or other particles with high energy to damage cancer cells [23]. After surgery, it is common to prescribe radiation therapy to avert cancer from coming back (i.e., recurrence). Other treatment options involve systematic therapy which comprises immunotherapy, targeted therapy, and chemotherapy [112].
2.3 Non-melanoma skin cancer
When healthy cells of the skin mutate and grow out of control, a tumor mass is formed [132]. Non-melanoma skin cancer can be partitioned into three main types (i.e., “Basal cells carcinomas,” “squamous cell cancer,” and “Merkel cell cancer”).
2.3.1 Non-Melanoma Development
Basal cells can be defined as the round-shaped cells existing in the lower epidermis. This cell type develops about \(80\%\) of non-melanoma cancer and is defined as “basal cell carcinomas” [103]. They mostly occur on the head and neck, and it is mainly produced by exposure to the sun. This type of skin cancer rarely expands to other body parts as it usually grows gradually. The epidermis is mostly formed up of flat scale-shaped cells called squamous cells. Approximately \(20\%\) of skin cancers arise from these cells and are named “squamous cell carcinomas” [103]. Sun exposure is the main cause of it, so it is diagnosed in many skin regions. Also, skin that has been exposed to X-rays, burned, or damaged by chemicals can develop this type of carcinoma. The percentage of squamous cell carcinomas expand to other body parts range from \(2\%\) to \(5\%\). “Merkel cell cancer” is a fast-growing or highly aggressive cancer [96]. It initiates at hormone-producing cells just below the hair follicles and the skin. It is commonly discovered in the head and neck area. It is worth mentioning that the rare types are reported graphically in Figure 1.
2.3.2 Non-melanoma statistics
In the USA alone, some people are diagnosed with more than one skin cancer type, so 3.3 million people are estimated to be diagnosed with 5.4 million cases of basal and squamous cell carcinoma [103]. For several years, non-melanoma cancer cases have been increasing. The causes of this increase are longer life spans, increased sun exposure, and earlier detection of the disease. When compared to each other, basal cell carcinoma is more popular than squamous cell carcinoma. In recent years, the rate of deaths from these skin cancers has decreased. Every month, more than 5, 400 people worldwide die of non-melanoma skin cancer [49].
2.3.3 Non-melanoma risk factors
Similar to melanoma, indoor tanning, Ultraviolet light exposure, and people with light-colored skin are more likely to develop non-melanoma skin cancer [81]. The risk of getting non-melanoma skin cancers rises as getting older. Women are less potential to develop this type of cancer than men. Exposure to considerable amounts of certain chemicals such as coal tar, arsenic, and paraffin rises the chance of developing skin cancer [70]. People with smoking habits have more potential to grow squamous cell cancer, particularly on the lips.
2.3.4 Non-melanoma diagnosis and treatment
Since it is rare for non-melanoma cancer to expand, a biopsy is usually the only test required to analyze and acquire the stage of cancer [81]. As mentioned in Sect. 2.2.5, a biopsy is the small amount of tissue extracted for testing beneath a microscope. For non-melanoma skin cancer, surgery is the main treatment [81]. It involves extracting the cancerous part and surrounding skin. Other treatments include anti-cancer creams, freezing (i.e., cryotherapy), photodynamic therapy, and radiotherapy.
3 Related studies
Research in the domain of melanoma detection, segmentation, recognition is still ongoing. Many automated approaches and techniques have been proposed to assist in computer-aided diagnosis. Previous related studies can be categorized into two main classifications: machine learning (ML) and deep learning (DL) classification techniques.
3.1 Classical machine learning-based approaches
The classical ML algorithms are consisting of many steps, e.g., pre-processing, feature extraction and reduction, and classification [91]. The accuracy of classification is based on the extracted features, so feature extraction is a key step. There are two main types of the extracted features. They are high-level (i.e., local) and low-level (i.e., global) features [121].
In Pugazhenthi et al. [98], a gray-level co-occurrence matrix (GLCM) is employed to extract the texture features, e.g., contrast, entropy, energy, and inverse difference moment from the segmented images. Then, these features were utilized to recognize the skin disease and classify it as melanoma, leprosy, or eczema using decision trees. An accuracy of \(87\%\) was obtained.
Arivuselvam et al. [9] used a Fuzzy clustering algorithm and the features were extracted from the input images by GLCM and Gabor filter in which features such as size, color, and texture were extracted. The SVM classifier was then utilized to calculate the feature values of 1, 500 dataset images and classify them.
In Khan et al. [71], a Gaussian filter is utilized to take out the noise from the images of the skin lesion followed by segmenting out the lesion by using an enhanced K-mean clustering, and a unique hybrid super-feature vector is formed. For the classification, an SVM is applied. Their proposed approach was evaluated using the DERMIS dataset, which has 397 skin cancer images, 251 were nevus and 146 were melanoma. An accuracy of \(96\%\) was obtained.
Astorino et al. [10] proposed a multiple instance learning algorithm. It was applied on 160 clinical data images and divided into 80 melanomas and 80 nevi. Their methodology obtained an accuracy of \(92.50\%\), a sensitivity of \(97.50\%\), and specificity of \(87.50\%\).
Balaji et al. [22] used a dynamic graph cut algorithm to perform the skin lesion segmentation followed by a Naive Bayes classifier for skin disorder classification. Their proposed method was tested by ISIC 2017 dataset and they achieved an accuracy of \(94.3\%\) for benign cases, \(91.2\%\) for melanoma, and \(92.9\%\) for keratosis.
In Murugan et al. [90], the watershed segmentation method was implemented to perform the segmentation task. The resultant segments were subjected to feature extraction method in which the ABCD rule, GLCM, and the shape of the extracted features were utilized for classification. They used four types of classifiers including K-nearest neighbor (KNN), random forest, and SVM. An accuracy of \(89.43\%\), sensitivity of \(91.15\%\), and specificity of \(87.71\%\) were achieved.
İlkin et al. [65] used the SVM algorithm as a classifier that utilizes a Gaussian radial basis function which had been enhanced by the bacterial colony algorithm. The proposed model was trained and evaluated using two datasets, namely ISIC and PH2. AUC values of \(98\%\) and \(97\%\) were obtained for ISIC and PH2, respectively.
3.2 Deep learning-based approaches
In the late 1990s, a shift from fully human-designed systems to computer-trained systems was delivered. This had done using sample data, from which the vectors of the handcrafted feature were extracted [50]. The next step was to allow the computers to figure out how to extract the suitable features from the input to perform the required task. The concept of learning how to extract features automatically from input data is the essence of numerous DL algorithms.
In Adegun and Viriri [2], an improved encoder–decoder network with sub-networks linked through a skip connection series was utilized for feature extraction and learning. Their algorithm was evaluated on two public datasets, PH2 and international symposium on biomedical imaging (ISBI) 2017. For the ISBI 2017 dataset, the reported accuracy and dice coefficient were \(95\%\) and \(92\%\), respectively, whereas the reported accuracy and dice coefficient were \(95\%\) and \(93\%\) for the PH2 datasets.
Albahli et al. [5] used YOLOv4-DarkNet and active contour for localization and segmentation of melanoma. Their algorithm was evaluated on ISIC 2016 and 2018. The reported dice score and Jaccard coefficient were 1 and 0.989.
Shan et al. [113] proposed FC-DPN segmentation topology. It was constructed over a dual-path and fully convolutional network. For the modified ISIC 2017 challenge test dataset, their proposed method gained a Jaccard index and an average dice coefficient of \(80.02\%\) and \(88.13\%\), respectively, while a Jaccard index and an average dice coefficient of \(83.51\%\) and \(90.26\%\) were obtained for the PH2 dataset.
Junayed et al. [67] introduced a CNN-based model to categorize skin cancer. Initially, a dataset was collected and divided into four categories of skin cancer images. Then, augmentation techniques were applied to increase the dataset size. On the test phase, their proposed model received a \(95.98\%\) accuracy, exceeding the GoogleNet and the MobileNet model by \(1.76\%\) and \(1.12\%\) respectively.
Alheejawi et al. [6] suggested a DL-based technique to segment regions of melanoma. Results obtained using a small dataset of melanoma images showed that the suggested approach could perform the segmentation with a dice coefficient around \(85\%\). Their method is proper for clinical examination as it had a short execution time with a fast turnaround time.
Vani et al. [128] suggested a DL-based system to predict the existence and the type of melanoma. For improving the images used for classification, pre-processing methods were utilized. CNN and self-organizing map (SOM) classifiers were utilized for the process of classification of melanoma. Their proposed system reported accuracy of \(90\%\) and specificity of \(99\%\).
Li and Jimenez [78] proposed a novel testing method based on the extreme learning machine network and AlexNet. Additionally, a new improved version of the Grasshopper optimization algorithm (GOA) was utilized to tune the hyperparameters of the proposed method. Their method was evaluated using the PH2 dataset with an accuracy of 98% and sensitivity of 93% and had the highest efficiency when compared to some different SOTA methods.
In Hasan et al. [56], an automated skin lesion classification framework was proposed. Their proposed method had merged the pre-processing and hybrid convolutional neural network. It had three distinct feature extractor modules that were fused to perform better-depth feature maps of the lesion. Lesion segmentation, augmentation, and class rebalancing were used to conduct the pre-processing phase. Three datasets named ISIC-2016, ISIC-2017, and ISIC-2018 datasets were utilized to evaluate the proposed algorithm. It had achieved an AUC of 96%, 95%, and 97%, for the three used datasets, respectively.
Maniraj and Maran [82] suggested a hybrid deep learning approach that utilized subband fusion of 3D wavelets. Their method consists of three stages (i.e., simple median filtering, 3D wavelet transform, and multiclass classification). The performance results on the PH2 dataset showed that it could effectively distinguish normal, benign, and malignant skin images with 99.33% average accuracy and more than 90% sensitivity and specificity.
3.3 Related studies summary
Table 1 summarizes the discussed related studies. They are organized in descending order concerning the publication year.
3.4 Plan of solution
In medical imaging applications, skin cancer detection, classification, and segmentation are important and difficult tasks. In the current study, various DL architectures are proposed to solve the skin cancer classification and segmentation problem. The transfer learning (TL) and SpaSA are used to tune (i.e., optimize) the training parameters and hyperparameters. Different experiments are performed and various performance metrics are utilized for evaluation. The best architectures are documented, stored, and reported to be used in further times.
4 Preliminaries
The current section discusses, for the reader, the background and elementary parts behind the proposed approach. The methodology section depends on them. It is organized into the following points:
-
Data scaling and augmentation.
-
Segmentation.
-
Deep learning (DL) classification.
-
Transfer learning (TL).
-
Parameters optimization.
-
Meta-heuristic optimization.
-
Performance metrics
4.1 Data scaling and augmentation
4.1.1 Data scaling
To normalize the scale of features or independent variables of data, scaling methods are employed. In data processing, scaling is generally conducted during the data pre-processing step to fit the data within a specific range [4, 88]. The four applied scaling techniques in the current research are (1) standardization, (2) normalization, (3) min-max scaling, and (4) max-absolute scaling.
-
Standardization: The standardization (i.e., z-score normalization) modifies the data, so the distribution has a mean and a standard deviation of 0 and 1 respectively.
-
Normalization: The dataset is re-scaled from its original range so that all values are in a new range [0 : 1].
-
Min-max scaling: In min-max scaling, the data are transformed so that the features are within a specified range.
-
Max-absolute scaling: The max-absolute scaling is obtained by finding the absolute maximum value in the dataset and dividing all the values in the column by that maximum value.
4.1.2 Data augmentation
Image data augmentation is a procedure that is employed to boost the dataset size artificially by generating altered versions of the images [86]. Data augmentation assists the coping with the “not enough data” issue, prevents over-fitting, and advances the ability of the models to generalize [16]. The transformation matrix can be also used to get the coordinates of a point after applying the data augmentation method on an image. Image augmentation methods adopted in the current study experiments are (1) flipping, (2) rotation, (3) shifting, (4) shearing, (5) zooming, (6) cropping, (7) color change, and (8) brightness change.
-
Flipping: Images can be flipped horizontally and vertically. In some frameworks, functions for vertical flips are not provided. Instead, a vertical flip can be employed by rotating an image by 180 degrees and then executing a horizontal flip.
-
Rotation: Rotation is accomplished by rotating the image on an axis between \({1^\circ }\) and \({359^\circ }\), rotating the image around the center or any other point, counterclockwise or clockwise. As the degree of rotation increases, data labels may be no longer preserved.
-
Shifting: Shifting the entire pixels of an image from one position to another position is known as shift augmentation. Two types of shifting (i.e., horizontal-axis and vertical-axis shift augmentation) exist.
-
Shearing: Shearing is used for shifting one part of the image like a parallelogram and transforming the orientation of the image.
-
Zooming: Zooming is applied to create images with varying levels of zooming. This augmentation zooms the image and adds new pixels for the image randomly. The image can be zoomed out or zoomed in.
-
Cropping: Random cropping is the method of cropping a part of the image, randomly. Similarly, center cropping is also employed to crop the image and applied when the image center holds more information than the corner.
-
Color changing: Data of digital images are regularly encoded as a tensor that has dimensions of \((\text {height} \times \text {width} \times \text {color channels})\). Color augmentation changes the pixel values instead of the position.
-
Brightness changing: Changing the brightness of the image is one way of performing data augmentation. Compared to the original image, the resultant one becomes lighter or darker.
4.2 Segmentation
Skin cancer segmentation algorithms are broadly categorized as thresholding, region-based, or edge-based methods [117]. Thresholding uses a combination of clustering, adaptive thresholding, and global thresholding. Good results can be achieved by thresholding methods when the contrast between the skin and the lesion is good. Hence, when the corresponding histogram is bimodal, but when the two regions overlap, it fails [100]. Edge-based methods function badly when the edges are not defined well, for example, when a smooth transmission between the skin and the lesion takes place. In these situations, the outline may leak within the edges as they have gaps. Region-based approaches face problems when the lesion area is structured or differently colored, leading to over-segmentation [68].
4.2.1 Threshold-based segmentation
Threshold-, pixel-, or point-based segmentation [64] is the simplest method to drive the segmentation of images, relying on grayscale values, to segment pixels in an image. Various algorithms have been suggested for skin segmentation and classification, including histogram-based thresholding and piecewise linear classifiers.
4.2.2 Edge-based segmentation
Boundary- or edge-based segmentation algorithms [68] usually mention dividing an image utilizing the boundaries between regions, by seeking border pixels and joining them to produce contours of the image. Nevertheless, for applying these methods manually and automatically, procedures are established. For manual methods, the mouse is used to lay lines that describe the edges of an image among regions, while for the automatic ones, some edge-detection filters are executed to divide the pixels into non-edge or edge based on the result of the filter output. Edge-detection filters include the Watershed segmentation algorithm, Laplacian of Gaussian filter, and Canny Edge Detector [130].
4.2.3 Region-based segmentation
In region-based segmentation methods, an image is segmented into groups of regions (i.e., similar pixels) relying on some features [69]. The core principle relies on the concept that inside the same area neighboring pixels have the same value. It can be achieved by comparing all pixels with their neighbors in a specific region and based on the similarity condition, the pixel is added to a particular region [130]. In the segmentation process, instead of the original input image, a featured image is used. The featured image is described with small neighborhoods from regions [119]. To use a region-based segmentation method, suitable threshold approaches have to be employed [68], as the noise has a significant influence on the output [130]. Some region-based methods are region splitting, region growing, and region merging.
4.2.4 Deep learning (DL) segmentation
DL-based image segmentation techniques can be assorted into: semantic, instance, panoptic, and depth segmentation sorts according to the segmentation goal. However, due to the huge variety in those tasks in terms of volume of work, the architectural categorization is used instead. The architectural grouping of these models includes CNNs [33], recurrent neural networks and long short term memory networks [60], encoder-decoders [11], and generative adversarial networks [52].
The U-Net model: U-Net [104] is an architecture used for semantic segmentation and is characterized by the symmetric U-shape. U-Net composes of an encoder and a decoder. The contracting path (i.e., encoder) is used to capture and collect context and the symmetric expanding path (i.e., decoder) is employed to enable accurate localization. The encoder obeys the typical architecture of a convolutional network. It is applied to transform the input volume into lower-dimensional space. The encoder has a modular structure composed of repeating convolution blocks. In the expansive path, every step is consisting of an up-sampling of the feature map. The decoder also has a modular structure, but its goal is to increase the spatial dimensions by reducing the encoder feature map.
The U-Net++ model: U-Net++ [140] architecture which can be considered as an extension of U-Net is essentially an encoder-decoder network that is deeply supervised where the sub-networks of the decoder and encoder are linked through a series of dense and nested skip paths. These re-designed paths seek to decrease the semantic gap between the sub-networks of the encoder and decoder. The U-Net++ architecture maintains the benefits of catching fine-grained details, producing better results of segmentation than U-Net.
The Attention U-Net model: Similar to the U-Net model, the Attention U-Net [93] includes expansion path at the right and contraction path at the left. At each level, it has a skip connection which is an attention gate. The attention gates are merged into the typical U-Net architecture to accentuate salient features that are pushed through the skip connections. For each skip connection, the gating signal aggregates information from several imaging scales which helps reach better performance and improves the resolution of the attention weights.
The V-Net model: The architecture of the V-Net [87] is very close to the widely used U-Net model, despite some differences. In the V-Net architecture, the left part can be separated into different phases running at various resolutions. In each stage, there are one to three convolution layers. To facilitate learning the residual function, the nonlinearities are used to process the input. Then, the processed input is employed in the convolution layers and appended to the output of the convolution layer of that phase. When compared to non-residual learning architecture such as U-Net, convergence is guaranteed by the V-Net network.
The Swin U-Net model: Swin-U-Net [30] is a U-shaped transformer-based architecture with skip connections for local and global feature learning. For an encoder, to extract context features, a hierarchical Swin transformer with shifted windows is employed. For the decoder, an asymmetric Swin Transformer with a patch expanding layer performs the up-sampling operation to restore the feature map’s spatial resolution.
4.3 Deep learning (DL) classification
The procedure of sorting a given data set into classes is known as classification. Classification can be done on structured and unstructured data. Its main goal is to map input variables to discrete output variables to identify which class the new data will fall into. Usually, the classes are mentioned as categories, targets, or labels. Several DL algorithms can be used to perform the task of classification such as CNNs [75], recurrent neural networks [111], long short-term memory networks [60], generative adversarial networks [52], radial basis function networks, [27], deep belief networks [59], and autoencoders [107]. In the current study, only CNN models are used to perform the classification.
4.3.1 Convolution neural network (CNN)
Neural networks are at the core of DL algorithms and are considered as an ML subset [51]. They are composed of layers of nodes, including an input layer, one or more hidden layers, and an output layer. Each node inside each layer has an associated weight and threshold and is connected to other nodes. If the node output is higher than the value of the threshold value, the node is triggered and starts to send data to the subsequent layer. Else, no data will be sent [32].
Neural networks are categorized into different types that are used to perform different tasks. For an instance, recurrent neural networks [111] are generally used for speech recognition and natural language processing while CNNs [73, 137] are frequently utilized for the tasks of computer vision and classification. Before the use of CNNs, to recognize objects in images, manual feature extraction methods. Now, CNNs offer a scalable approach to recognize and classify images. For the training process, they need graphical processing units (GPUs) so they are computationally demanding [76].
CNNs differ from other types by their higher performance with image, audio signal, or speech input types [12]. They consist of three primary layers types, namely convolutional, pooling, and fully connected (FC) layers. The convolution layer is the earliest layer of a typical CNN. Following it, additional convolution and pooling layers exist, and the final layer is an FC one [14]. After each layer, the complexity of the CNN increases and identifies larger parts of the image. As the image proceeds through the CNN layers, more extensive elements of the object are begun to be recognized until the expected object is finally recognized [53].
Convolution layer: The central building block of a CNN is the convolutional layer as the computation majority happens inside it. However, it requires some elements, e.g., input data, a feature map, and a filter. A tensor, also called a kernel or filter, seeks for the presence of the features, which is known as a convolution [39]. The kernel is a 2D weights array that symbolizes an image part. During training, back-propagation and gradient descent are used to adjust parameters like weight values. Yet, the three hyperparameters that affect the size of the output and have to be adjusted before the training begins include [79]:
-
The number of filters: The output depth is affected by it. For illustration, three different feature maps associated with three distinct depths are produced by three different filters.
-
Stride: The pixels number that the kernel proceeds through the input matrix is defined as stride.
-
Padding: Discussed in the following paragraph.
There are different types of padding [7]:
-
Zero padding: It is typically used when the filters and the input image do not meet. All elements that lay outside the input matrix are set to zero, so a greater or equal output is produced.
-
Valid padding: Also, it is named as no padding. In this type, if the dimensions do not match, the last convolution will be discarded.
-
Same padding: This type guarantees that the output and input layers are of the same size.
-
Full padding: In this type, the size of the output is increased by appending zeros to the input border.
Activation function: The linear convolution outputs are passed into a nonlinear activation function. Before, the used nonlinear functions are the smooth ones such as tangent hyperbolic (i.e., Tanh) or Sigmoid functions [99]. Lately, the most widely used function is the rectified linear unit (ReLU) [21]. ReLU is a piecewise linear function that returns 0 if a negative input is received, else, it returns the input value. Thus the output range from 0 to infinity. For neural network types, it has evolved as the standard activation function. As, its architecture is simpler, easier to train, and often outperforms others [28].
Pooling layer: The pooling layers (i.e., down-sampling layers) are used to perform dimensionality reduction and minimize the number of input parameters. Similar to the convolution one, a filter is passed over the entire input by the pooling operations. The distinction is that the filter contains no weights. Rather, a summation function is applied by the kernel to the values in the receptive field, populating the output array [31]. The major pooling types are:
-
Max pooling: The input is passed to a filter that specifies the pixel holds the maximum value to be sent to the output array, in which (1) patches are extracted from the feature maps of the input, (2) in each patch, the maximum value is generated, and (3) the other values are discarded [110].
-
Average pooling: The input is passed to a filter that calculates the average value to be sent to the output array. In the pooling layer, a bunch of information is lost, but several benefits are gained. They assist to lower the CNN complexity, enhance efficiency, and restrict the risk of over-fitting [124].
Fully connected (FC) layer: Simply, an FC Layer is a feed-forward neural network. The few last layers in the network are FC layers. The output of the last convolution or pooling layer (i.e., feature maps) is regularly flattened and then fed into the FC layer. A learnable weight is utilized to connect every input to every output [19]. The final layer usually has several output nodes same as the number of classes. Each FC layer is followed by a nonlinear function such as ReLU [15].
4.4 Transfer learning (TL)
The reuse of a formerly trained model for a novel problem is called transfer learning (TL). In DL, it becomes popular to use TL as deep neural networks that can be trained with relatively small data. It becomes very helpful in data science, as the majority of the real-world problems do not have a considerable amount of classified data to train complex models [18, 126]. In TL, what has been learned in one task is exploited to enhance the second task generalization. Generalization can be done by loosening the assumption that the test and training data must be identically distributed and independent [13]. The extensive idea is to employ the knowledge that has been gained by the model trained with plenty of labeled data in a novel task with small amount of data [94]. There are five types of transfer learning which includes (1) domain adaptation, (2) domain confusion, (3) multitask learning, (4) one-shot learning, and (5) zero-shot learning [109].
The TL process is divided into four contexts relying on “what to transfer” in learning. They involve approaches of (1) the instance-transfer, (2) the feature-representation-transfer, (3) the parameter-transfer, and (4) the relational-knowledge-transfer [1]. TL uses a previously trained stored model as a starting point for DL. This enables fast progress and improved performance [80]. Many pre-trained CNN models are available to be used such as VGG16 [118], ResNet [58], MobileNet [62], Xception [34], NASNet [141], and DenseNet [63].
NASNet, VGG, and MobileNet architectures are utilized for image classification. The used architectures in the current study are NASNetLarge and NASNetMobile [141], MobileNet [62], MobileNetV2 [108], MobileNetV3Small and MobileNetV3Large [61], and VGG16 and VGG19 [118]. In all experiments of classification, the size of the input image is set to \((100 \times 100 \times 3)\).
4.4.1 NASNet
Google ML group has materialized the idea of an optimized network through the concept of NAS that is based on reinforcement learning [141]. The architecture is consisted of a controller RNN and CNN, which are to be trained. The NASNet is trained with two sizes of input images of \(331 \times 331\) and \(224 \times 224\) to obtain NASNetLarge and NASNetMobile architectures respectively. When moving from NASNetMobile to NASNetLarge, there is great growth in several parameters. NASNetMobile and NASNetLarge have 5, 326, 716 and 88, 949, 818 parameters, respectively, which makes NASNetLarge less reliable.
4.4.2 MobileNet
MobileNet is created to efficiently increase accuracy whereas being aware of the limited resources for an on-device. To meet the resource constraints of the computing devices, MobileNet has low-latency and low-power models [61]. MobileNet uses separable filters, which is a mixture of a point- and a depth-wise convolution. It operates filters with a size of \(1 \times 1\) for minimizing the normal convolution operation computational overheads. Hence, the network is lighter in terms of size and computational complexity. The MobileNet has 4.2 million parameters with input image of size \(224 \times 224 \times 3\).
4.4.3 MobileNetV2
MobileNetV2 architecture is close to the original MobileNet, except that inverted residual blocks with bottlenecking features were utilized and nonlinearities in narrow layers were removed [108]. It has fewer parameters than the original MobileNet. MobileNets support all input sizes larger than \(32 \times 32\) with greater image sizes giving a better performance. In MobileNetV2, two sorts of blocks exist. The first is a 1 stride-sized residual block. The other is used for downsizing and it is a 2 stride-sized block. For both types of blocks, there are 3 layers. The initial layer is 1\(\times\)1 convolution with ReLU6, the next is a depth-wise convolution, and the last one is a further \(1 \times 1\) convolution but without any activation function.
4.4.4 MobileNetV3
The major contribution of MobileNetV3 is the utilizing of AutoML to obtain the best possible neural network architecture for a given problem. Precisely, MobileNetV3 combines pair of AutoML techniques: NetAdapt and MnasNet [61]. MobileNetV3 initially uses MnasNet to search for a coarse architecture. MnasNet utilized reinforcement learning to select the optimal configuration from a discrete set of choices. Then, the architecture is fine-tuned using NetAdapt which cuts the underutilized activation channels in small increments. MobileNetV3 is represented as two models: MobileNetV3Large and MobileNetV3Small are targeted at high and low resource use cases, respectively. When compared to MobileNetV2, MobileNetV3Large is \(3.2\%\) more accurate on ImageNet classification and latency is decreased by \(20\%\). Similarly, MobileNetV3Small is more accurate by \(6.6\%\) with comparable latency.
4.4.5 VGG model
The VGG is formed up of convolution and pooling layers stacked together [118]. In VGG16, the network depth is 16 layers without the issue of vanishing gradients. It includes 13 convolution layers, 5 max-pooling layers, and 3 dense layers with two 4096 sized layers. A nonlinear ReLU activation function was used by all the hidden layers while the final layer uses a SoftMax function. On the contrary, the VGG19 network depth is 19 layers. There are 16 convolution layers, 5 MaxPool layers, and 3 dense layers with two 4096 sized layers. Similar to VGG16, all the hidden layers of VGG19 utilize the ReLU activation function and the final layer uses the SoftMax function.
Table 2 summarizes the classification models used in the current study.
4.5 Parameters optimization
Parameters optimization is the process of selecting the values of parameters that are optimal for some desired purpose (e.g., minimizing an error function). The parameters are the weights and biases of the network. The cost (i.e., error) function is used to perform the model predictions and target values comparison [129]. Some of the weights optimizers are gradient descent algorithm [106], Adam (adaptive moment optimization algorithm) [72], Nadam [123], Adagrad [89, 131], AdaDelta [136], AdaMax [40], and Ftrl [85].
To minimize the error function, the gradient descent algorithm [106] updates the parameters. Small steps in the negative direction of the loss function are taken by this algorithm. Adam [72] merges the heuristics of the RMSProp [42] and momentum [134] optimization algorithms. The momentum optimizer speeds up the search in the minima direction, while the RMSProp optimizer prevents the search in the direction of the oscillation.
4.6 Meta-heuristic optimization
Usually, numerous real-world optimization problems involve a big number of decision variables, complex nonlinear constraints, and objective functions; hence, they are increasingly becoming challenging. When the objective constraints have multi-peaks, the traditional optimization approaches such as numerical methods become less powerful. Meta-heuristic optimization methods become powerful tools for managing optimization issues. Their popularity drives by the following aspects [139]:
-
Simplicity: These meta-heuristic methods are mathematical models derived from nature and are generally simple, easy to perform, and develop variants according to existing approaches.
-
Black box: For a given problem, a set of inputs can offer a set of outputs.
-
Randomness: This allows the meta-heuristic algorithm to prevent trapping into local optima and inspect the entire search space.
-
Highly flexible: Their practicality can be implied to diverse types of optimization problems, e.g., complex numerical problems with plentiful local minima, nonlinear problems, or non-differentiable problems.
4.6.1 Sparrow search algorithm (SpaSA)
The sparrow search algorithm (SpaSA) [135] is inspired by the strategies of foraging and the behaviors of anti-predation of sparrows. Compared with traditional heuristic search methods, it has strong optimization ability, fast convergence speed, and more extensive application procedures. Hence, the SpaSA is captivating the attention of researchers in various fields.
It was originally suggested by Xue and Shen [135]. The sparrow population is divided into (1) the discoverer and (2) the follower sparrows according to their role in the food search procedure. Each of them does their behavioral strategies separately. In most cases, the discoverers are 0.2 of the population size. They are the guiders, leading other individuals in the food search. To obtain more food, the roles are switched flexibly between the discoverers and the followers and compete for the food resources of their companions. However, the proportion of the followers and the discoverers inside the population is fixed. The individuals’ energy and the sparrows’ anti-predation behavior determine their foraging strategies. The mathematical representation of the SpaSA algorithm will be discussed in Sect. 5.4.1.
Why the sparrow search algorithm (SpaSA) has been selected to be used? SpaSA is a relatively new swarm intelligence heuristic algorithm. As reported in [135], results showed that the proposed SpaSA is superior to grey wolf optimizer (GWO), gravitational search algorithm (GSA), and particle swarm optimization (PSO) in terms of accuracy, convergence speed, stability, and robustness. Additionally, the SpaSA has high performance in diverse search spaces. Using the SpaSA, the local optimum issue is avoided effectively as it has a good ability to explore the potential region of the global optimum.
4.7 Performance metrics
Evaluating the quality of the produced output is accomplished by comparing images is an essential part of measuring progress [138]. Performance metrics are distinct from loss functions. Loss functions give a measure of model performance. Metrics are employed to estimate the performance of a model. Nevertheless, the loss function can also be utilized as a performance metric. The assessment metric should provide details related to the task, whether it is interventional or diagnostic. For illustration, some tasks require real-time operations, while tasks for diagnostic procedures can be conducted offline. For choosing the optimal approach, the importance of different performance metrics may differ. Performance metrics can be categorized as (1) spatial overlap-based metrics, (2) probabilistic-based metrics, (3) pair-counting-based metrics, (4) volume- or area-based metrics, (5) information theoretic-based metrics, and (6) spatial distance-based metrics [120]. In the current study, only spatial overlap- and probabilistic-based metrics were used, so they will be discussed in this section.
4.7.1 Spatial overlap-based metrics
The overlap-based performance metrics are the ones that can be acquired from the cardinalities of the confusion matrix. A confusion matrix is a matrix created to assess the model performance. It matches the actual values with the ones predicted by the model. It consists of: (1) true positive (TP), (2) true negative (TN), (3) false positive (FP), and (4) false negative (FN).
The accuracy is the ratio of the correct predictions for the test data to the total predictions. The true negative rate (TNR), also termed specificity, estimates the ability of the model to predict the true negatives of every class. Likewise, the true positive rate (TPR), also termed sensitivity or recall, is defined as the ratio of samples that were predicted to belong to a class to all of the samples that truly belong to this class. Hence, it estimates the model’s ability to predict the true positives of every class.
The false negative rate (FNR) and the false positive rate (FPR) (i.e., fallout) are another two metrics associated with the two previously mentioned metrics. Additionally, precision also termed positive predictive value (PPV) is the ratio of true positives among the retrieved instances.
The dice coefficient, also termed as the overlap index and F1-score, is one of the most applied metrics to evaluate the medical images [46]. Besides the direct comparison between the true and predicted value, it is often used to estimate repeatability. The Jaccard index (JAC), also termed as the intersection over union (IoU), is the intersection among two sets divided by their union [66].
4.7.2 Probabilistic-based metrics
Probabilistic-based metrics are defined as a statistical function’s measure estimated from the voxels in the overlap region. The receiver operating characteristic (ROC) curve is a relationship graph between FPR and TPR. The area under the ROC curve (AUC) was suggested by Hanley and McNeil [55] as an evaluation of the accuracy of diagnostic radiology In the situation of comparing the predicted and true value, the AUC defined according to [97] is considered, that is, the trapezoidal area determined by the lines \(\text {TPR} = 0\) and \(\text {FPR} = 1\) and the measurement point.
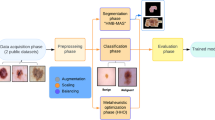
5 Methodology and suggested approach
In summary, the images is accepted by the input layer. In the next phase, they are pre-processed by employing dataset augmentation, scaling, and balancing. The images can be classified and segmented after that using the suggested pre-trained models. Finally, the transfer learning and meta-heuristic optimization phase occurs. After completion, the figures, statistics, and post-trained models are prefaced. In the following subsections, these phases are debated.
5.1 Dataset acquisition
In the current study, five publicly available datasets are utilized and downloaded from Kaggle. The first dataset is named “ISIC 2019 and 2020 Melanoma dataset” [37, 38, 105, 127]. It is composed of 11,449 images. It is partitioned into 2 classes: “MEL” and “NEVUS.” It can be downloaded and used from https://www.kaggle.com/qikangdeng/isic-2019-and-2020-melanoma-dataset.
The second one is named “Melanoma Classification (HAM10000)” [36, 127]. It is composed of 10,015 images in which images have different sizes. It is partitioned into 2 classes: “Melanoma” and “NotMelanoma.” It can be downloaded and used from https://www.kaggle.com/adacslicml/melanoma-classification-ham10k.
The third one is named “Skin diseases image dataset”. It is composed of 27, 153 images in which images have different sizes. It is partitioned into 10 classes: “Atopic Dermatitis,” “Basal Cell Carcinoma,” “Benign Keratosis-like Lesions,” “Eczema,” “Melanocytic Nevi,” “Melanoma,” “Psoriasis pictures Lichen Planus and related diseases,” “Seborrheic Keratoses and other Benign Tumors,” “Tinea Ringworm Candidiasis and other Fungal Infections” and “Warts Molluscum and other Viral Infections.” It can be downloaded and used from https://www.kaggle.com/ismailpromus/skin-diseases-image-dataset.
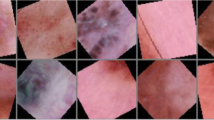
The fourth one is named “Skin cancer segmentation and classification”. It is composed of 10, 015 images in which images are of different sizes. It can be downloaded and used from https://www.kaggle.com/surajghuwalewala/ham1000-segmentation-and-classification. The fifth one is named “PH2”. It is composed of 200 dermoscopic images of melanocytic lesions. It can be downloaded and used from https://www.fc.up.pt/addi/ph2%20database.html.
Table 3 summarizes the used datasets, and Fig. 2 shows samples from them.
5.2 Dataset pre-processing
5.2.1 dataset scaling
Data scaling is discussed in Sect. 4.1.1 and the corresponding equations that are used in the current study are Eq. 1 for standardization, Eq. 2 for normalization, Eq. 3 for the min-max scaler, and Eq. 4 for the max-absolute scaler where \(\mu\) is the image mean and \(\sigma\) is the image standard deviation.
5.2.2 Dataset augmentation and balancing
Before the training process, data balancing is applied to balance the categories since the number of images per category is not even. Data balancing is performed using the methods of data augmentation discussed in Sect. 4.1.2. The used ranges in this process are (1) \(25^{\circ }\) for rotation, (2) \(15\%\) for shifting the width and height, (3) \(15\%\) for shearing, (4) applying horizontal and vertical flipping, and (5) changing the brightness in the range of [0.8 : 1.2]. Additionally, in the learning and optimization phase, data augmentation is used to augment the images to avoid any over-fitting and increase the diversity [17]. The used transformation metrics are Eq. 5 for horizontal flipping (i.e., x-axis), Eq. 6 for rotation, Eq. 7 for shifting, Eq. 8 for shearing, and Eq. 9 for zooming where \(\theta\) is the rotation angle, \(t_{x}\) determines the shifting along x-axis, while \(t_{y}\) determines the shifting along y-axis, \(sh_{x}\) determines the shear factor along x-axis while \(sh_{y}\) determines the shear factor along y-axis, and \(C_{x}\) determines the zoom factor along x-axis and \(C_{y}\) determines the zoom factor along y-axis.
5.3 Segmentation phase
The segmentation phase is qualified for segmenting the tumor portion from the medical skin images. In the current phase, the U-Net with different five flavors designed for image segmentation was used. The used U-Net models in the present study are U-Net [104], U-Net++ [140], Attention U-Net [93], V-Net [87], and Swin U-Net [30]. The U-Net consists of the left contraction path and the right expansion path. In the current study, there are four different configurations applied to three U-Net models (i.e., U-Net, U-Net++, and Attention U-Net). They are (1) the default left contraction path is used in two settings and it is replaced with the VGG19 and DenseNet121 architectures for the other two settings, (2) the pre-trained weights for the VGG19, DenseNet121 are set with ImageNet, (3) the ImageNet weights are frozen from being updated, (4) the depth of the architecture is set to five with the number of filters of [64, 128, 256, 512, 1024] in each level (i.e., block), (5) the input image size is set to \((128 \times 128 \times 3)\), and (6) the output mask size is \((128 \times 128 \times 1)\).
5.3.1 The U-Net model
In the current study, four configurations of U-Net are employed to perform the segmentation task. In the first configuration, the architecture provided in [104] is utilized. Whereas batch normalization and GeLU as a hidden activation function is applied in the second one. For the third and fourth configurations, in addition to batch normalization and GeLU hidden activation function, VGG19 and DenseNet201 are utilized as a network backbone (i.e., replace the encoder with the SOTA architectures). A summarization of the four different U-Net configurations is presented in Table 4.
5.3.2 The U-Net++ model
Similarly to U-Net, four configurations of U-Net++ are employed to perform the segmentation task. In the first configuration, the architecture provided in [140] is utilized, Whereas batch normalization and GeLU as a hidden activation function is applied in the second one. For the third and fourth configurations, in addition to batch normalization and GeLU hidden activation function, VGG19 and DenseNet201 are utilized as a backbone. For the four configuration, deep supervision is deactivated. A summarization of the four different U-Net++ configurations is presented in Table 5.
5.3.3 The Attention U-Net model
Similar to the previous two architectures, four configurations of Attention U-Net are used to perform the segmentation task. In the first configuration, the architecture provided in [93] is utilized, whereas the remaining configurations are the same as the previous two networks. For the four configuration, ReLU was used as an attention activation function and Add is used as an attention type. A summarization of the four different Attention U-Net configurations is presented in Table 6.
5.3.4 The V-Net model
In this study, only one configuration is used for the v-net model. GeLU is used as the hidden activation function, batch normalization is applied, and the pooling and un-pooling are deactivated. The configuration of the V-Net architectures is presented in Table 7.
5.3.5 The Swin U-Net model
Similar to V-Net, only one configuration is used for the swin u-net model. The configuration of the Swin U-Net architecture is presented in Table 8.
5.4 Learning and optimization
To achieve the SOTA performance, different DL training hyperparameters (as shown in Table 9) required to be optimized. Try-and-error, grid search, and meta-heuristic optimization algorithms are techniques used to optimize the hyperparameters. Try-and-error is a weak technique as it does not cover the ranges of the hyperparameters. However, the grid search covers it, but, to complete the searching process, a long time (e.g., months) is required.
In the current study, we are optimizing (1) loss function, (2) dropout, (3) batch size, (4) the parameters (i.e., weights) optimizer, (5) the pre-training TL model learn ratio, (6) the dataset scaling technique, (7) do we need to apply augmentation or not, (8) width shift range, (9) rotation range, (10) shear range, (11) height shift range, (12) horizontal flipping, (13) zoom range, (14) vertical flipping, and (15) brightness range. In the case the sixth hyperparameter is true, the last eight hyperparameters will be optimized; otherwise, they will be neglected. Hence, at least 6 hyperparameters are required to be optimized. So, if the grid search approach is utilized, this will lead to \(O(N^6)\) concerning the running complexity. As a result, the meta-heuristic optimization algorithms approach using Sparrow Search Algorithm (SpaSA) is applied in the current study.
5.4.1 Sparrow search algorithm (SpaSA)
This approach will be used to solve the optimization issue to obtain the best combinations. As a beginning, all sparrow populations and their parameters are initialized randomly from the given ranges (as described in Table 11). The steps of the hyperparameters optimization include (1) objective function calculation, (2) population sorting, (3) selection, and (4) updating. After operating a set of iterations, the best global optimal location and fitness value are reported. The steps are explained comprehensively in the next subsections.
5.4.2 Initial population
Initially, the sparrows population and its relevant parameters are selected randomly. An arbitrary method is used to generate the initial population in SpaSA. It can be defined as shown in Eq. 10 where \(\text {X}_{i,j}\) is the position of \(i^{th}\) sparrow in \(j^{th}\) search space, i is the solution index, and j is the dimension index. D in the current study will be set to 15 (i.e., the number of hyperparameters required to be optimized). It will generate a population with a size of \((Ps \times D)\) where Ps is the population size (i.e., number of sparrows) and its value is set to 10 in the current study.
5.4.3 Objective function calculation
The objective function is applied to each sparrow to determine the corresponding score. The current problem is a maximization one, the higher the value, the better the sparrow. To simplify this step, the objective function can be thought of as a black box in which the solution is the input and the score (i.e., accuracy in this case) will be output. What happens internally? After accepting the solution required to be evaluated, the 15 elements mentioned earlier are extracted and applied to the pre-trained CNN model (e.g., VGG16). Initially, the model uses these certain values to start the learning process (i.e., the training and validation processes). Then, it evaluates itself on the entire dataset to find the overall performance metrics. Finally, the objective function returns the accuracy. The reported performance metrics are discussed in Sect. 4.7 and their equations are Eq. 11 for accuracy, Eq. 12 for specificity, Eq. 13 for recall, Eq. 14 for FNR, Eq. 15 for fallout, Eq. 16 for precision, Eq. 17 for dice coef., Eq. 18 for JAC, and Eq. 19 for AUC.
5.4.4 Population sorting
After calculating the objective function of each sparrow in the population set, sparrows are sorted in descending arrangements concerning the values of the objective function.
5.4.5 Selection
The current best individual \(X_{\rm best}^{t}\) and worst individual \(X_{\rm worst}^{t}\) and their fitness values are picked to be applied in the updating process.
5.4.6 Population updating
Using SpaSA, the individual with the best fitness values has the priority to collect food in the search procedure and oversee the entire population movement. So, updating the sparrow location for producers is important and can be done using Equation 20 where h represents the number of the current iteration and T is the maximal iterations number. \(X_{i,j}\) represents the current position of the ith sparrow in the jth dimension. \(\alpha\) is a random number \(\in [0,1]\). Q is a random number from the normal distribution. L represents a \(1 \times D\) matrix containing all 1 element. R2 and ST represent warning and safety values respectively, and \(R2 \in [0,1]\), \(ST \in [0.5,1]\). When \(R2 < ST\), there are no predators and the discoverers can widely search for food sources. Otherwise, some sparrows have detected the predators and the whole population flies to other safe areas when the chirping alarm happens.
Additionally, some of the followers supervise the discoverers and those discoverers with high predation rates for food, increasing their nutrition. The followers’ position is updated using Equation 21 where \(X_P\) is the currently optimal discoverer position, and \(X_{\rm worst}\) indicates the current worst position. A is a \(1 \times D\) matrix, where an element is only − 1 or 1, with \(A^+ = A^{\rm T} \times (A \times A^T)^{-1}\). If \(i > 0.5 \times n\), when the followers are starving and have low levels of energy reserves, they leave to search for food in other areas. The movement of the leaving followers is in a random direction which is away from the current worst position. Otherwise, the followers with high levels of energy move to the discoverers that have found good food.
It is assumed that only \(10\%\) to \(20\%\) of the entire sparrow population are aware of the danger. The sparrows initial positions are randomly formed in the population using Eq. 22 where \(\beta\), the step size control parameter, is a normal distribution with a mean value of 0 and a variance of 1 of random numbers. \(X_{\rm best}\) is the current global optimal location. \(K \in [-1, 1]\) is a random number that denotes the direction in which the sparrow moves. \(\epsilon\) is the smallest constant to avoid zero-division-error. fi is the fitness value of the present sparrow; hence, fg and fw are the current global best and worst fitness values, respectively. When \(fi > fg\), it indicates that the sparrow is at the edge of the group. \(X_{\rm best}\) represents the location of the center of the population and is safe around it. While \(fi = fg\) reveals that the sparrows, that are in the middle of the population, are aware of the danger.
5.5 The overall pseudocode and flowchart
The steps are iteratively computed for a number of iterations. Algorithm 1 and the corresponding flowchart in Fig. 3 summarize the proposed learning and optimization approach.

6 Experiments and discussions
The experiments are divided into two categories: (1) segmentation experiments and (2) optimization, learning, and classification experiments.
6.1 Experiments configurations
Generally, “Python” programming language is used in the current study for coding and testing. Google Colab, with its GPU, is the learning and optimization environment. Tensorflow, Keras, keras-unet-collection, NumPy, OpenCV, Pandas, and Matplotlib are the major used Python packages [20]. The dataset split ratio is set to \(85\%\) (for training and validation) and \(15\%\) (for testing). Dataset shuffling is applied randomly during the learning process. The images are resized to \((100 \times 100 \times 3)\) for classification and to \((128 \times 128 \times 3)\) for segmentation in the RGB color space. Table 10 summarizes the common configurations of the experiments, Table 11 summarizes the optimization, learning, and classification specific configurations, and Table 12 summarizes the segmentation specific configurations.
6.2 Segmentation experiments
The current subsection presents and discusses the experiments related to segmentation. The experiments are applied using U-Net [104], U-Net++ [140], Attention U-Net [93], Swin U-Net [30], and V-Net [87]. Table 13 shows the summarization of the reported results related to the segmentations experiments. For the “Skin cancer segmentation and classification” dataset, Table 13 shows that the best model is the “U-Net++-DenseNet201” concerning the loss, accuracy, F1, AUC, IoU, and dice values. However, the “U-Net++-Default” model is the best concerning the specificity, hinge, and square hinge values. It worth mentioning that the “V-Net-VGG19” model is better than other concerning the sensitivity and recall values and the “Attention U-Net-DenseNet201” models is better concerning the precision value. Figure 4 presents a graphical summarization of the reported segmentation results concerning the “Skin cancer segmentation and classification” dataset. For the “PH2” dataset, Table 14 shows that the best model is the “Attention U-Net-DenseNet201” concerning the loss, accuracy, F1, IoU, and dice values. However, the “Swin U-Net” model is the best concerning the precision, specificity, and squared hinge values. It worth mentioning that the “UNet++-Default” model is ignored as it reported meaningless performance metrics. Figure 5 presents a graphical summarization of the reported segmentation results concerning the “PH2” dataset.
6.3 Learning and optimization experiments
The current subsection presents and discusses the experiments related to the learning and optimization experiments using the mentioned pre-trained TL CNN models (i.e., MobileNet, MobileNetV2, MobileNetV3Small, MobileNetV3Large, VGG16, VGG19, NASNetMobile, and NASNetLarge) and SpaSA meta-heuristic optimizer. The number of epochs is set to 5. The numbers of SpaSA iterations and population size are set to 10 each. The captured and reported metrics are the loss, accuracy, F1, precision, recall, sensitivity, specificity, AUC, IoU coef., Dice coef., cosine similarity, TP, TN, FP, FN, logcosh error, mean absolute error, mean IoU, mean squared error, mean squared logarithmic error, and root mean squared error.
6.4 The “ISIC 2019 and 2020 Melanoma dataset” experiments
Table 15 shows the TP, TN, FP, and FN of the best solutions after the learning and optimization processes on each pre-trained model concerning the “ISIC 2019 and 2020 Melanoma dataset” dataset. It shows that MobileNet pre-trained model has the lowest FP and FN values. On the other hand, MobileNetV3Small has the highest FP and FN values. The best solutions combinations concerning each model are reported in Table 16. It shows that the KLDivergence loss is recommended by five models while the Poisson loss is recommended by two models only. The AdaGrad parameters optimizer is recommended by four models, while the SGD Nesterov parameters optimizer is recommended by two only. All models recommended applying data augmentation. The min-max scaler is recommended by three models. From the values reported in Table 15 and the learning history, we can report different performance metrics. The reported metrics are partitioned into two types. The first reflects the metrics that are required to be maximized (i.e., Accuracy, F1, Precision, Recall, Specificity, Sensitivity, AUC, IoU, Dice, and Cosine Similarity). The second reflects the metrics that are required to be minimized (i.e., Categorical Crossentropy, KLDivergence, Categorical Hinge, Hinge, SquaredHinge, Poisson, Logcosh Error, Mean Absolute Error, Mean IoU, Mean Squared Error, Mean Squared Logarithmic Error, and Root Mean Squared Error). The first category metrics are reported in Table 17, while the second is in Table 18. From them, we can report that the MobileNet pre-trained model is the best model compared to others concerning the “ISIC 2019 and 2020 Melanoma dataset” dataset. Figure 6 and Figure 7 present graphical summarizations of the reported learning and optimization results concerning the “ISIC 2019 and 2020 Melanoma dataset” dataset.
6.5 The “Melanoma Classification (HAM10K)” experiments
Table 19 shows the TP, TN, FP, and FN of the best solutions after the learning and optimization processes on each pre-trained model concerning the “Melanoma Classification (HAM10K)” dataset. It shows that MobileNet pre-trained model has the lowest FP and FN values. On the other hand, MobileNetV3Small has the highest FP and FN values. The best solutions combinations concerning each model are reported in Table 20. It shows that the KLDivergence loss is recommended by four models while the Squared Hinge loss is recommended by two models only. The SGD Nesterov parameters optimizer is recommended by five models while the SGD parameters optimizer is recommended by two only. Seven models recommended applying data augmentation. The min-max scaler is recommended by four models. From the values reported in Table 19 and the learning history, we can report different performance metrics. The reported metrics are partitioned into two types. The first reflects the metrics that are required to be maximized (i.e., Accuracy, F1, Precision, Recall, Specificity, Sensitivity, AUC, IoU, Dice, and Cosine Similarity). The second reflects the metrics that are required to be minimized (i.e., Categorical Crossentropy, KLDivergence, Categorical Hinge, Hinge, SquaredHinge, Poisson, Logcosh Error, Mean Absolute Error, Mean IoU, Mean Squared Error, Mean Squared Logarithmic Error, and Root Mean Squared Error). The first category metrics are reported in Table 21, while the second is in Table 22. From them, we can report that the MobileNet pre-trained model is the best model compared to others concerning the “Melanoma Classification (HAM10K)” dataset. Figure 8 and Figure 9 present graphical summarizations of the reported learning and optimization results concerning the “Melanoma Classification (HAM10K)” dataset.
6.6 The “Skin diseases image dataset” experiments
Table 23 shows the TP, TN, FP, and FN of the best solutions after the learning and optimization processes on each pre-trained model concerning the “Skin diseases image dataset.” It shows that MobileNetV2 pre-trained model has the lowest FP and FN values. On the other hand, MobileNetV3Small has the highest FP and FN values. The best solutions combinations concerning each model are reported in Table 24. It shows that the KLDivergence loss is recommended by five models, while the Categorical Crossentropy loss is recommended by three models only. The AdaMax parameters optimizer is recommended by four models, while the SGD parameters optimizer is recommended by two only. Seven models recommended applying data augmentation. The standardization is recommended by four models. From the values reported in Table 23 and the learning history, we can report different performance metrics. The reported metrics are partitioned into two types. The first reflects the metrics that are required to be maximized (i.e., Accuracy, F1, precision, recall, specificity, sensitivity, AUC, IoU, dice, and cosine similarity). The second reflects the metrics that are required to be minimized (i.e., Categorical crossentropy, KLDivergence, categorical hinge, hinge, squaredhinge, poisson, logcosh error, mean absolute error, mean IoU, mean squared error, mean squared logarithmic error, and root mean squared error). The first category metrics are reported in Table 25, while the second is in Table 26. From them, we can report that the MobileNetV2 pre-trained model is the best model compared to others concerning the “Skin diseases image dataset.” Figures 10 and 11 present graphical summary of the reported learning and optimization results concerning the “Skin diseases image dataset.”
6.7 Overall discussions
The experiments conducted in this study are split into two categories, i.e., segmentation and classification. In the segmentation experiments, different U-Net models were used. Concerning the “Skin cancer segmentation and classification” dataset, the best model was the U-Net++ with DenseNet201 as a backbone regarding the loss, accuracy, F1-score, AUC, IoU, and Dice values. However, the Attention U-Net model was the best regarding the AUC value. The worst results were obtained by the default UNet++ architecture. The achieved scores by U-Net++ with DenseNet201 as a backbone architecture were \(94.16\%\), \(91.39\%\), \(99.03\%\), \(96.08\%\), \(96.41\%\), \(77.19\%\), \(75.47\%\) in terms of accuracy, F1-score, AUC, IoU, Dice, hinge, and squared hinge. Concerning the “PH2” dataset, the best model was the Attention U-Net with DenseNet201 as a backbone regarding the loss, accuracy, F1, IoU, and dice values. However, the “Swin U-Net” model is the best concerning the precision, specificity, and squared hinge values. The achieved scores by Attention U-Net with DenseNet201 as a backbone architecture were 0.137, 94.75%, 92.65%, \(92.56\%\), \(92.74\%\), \(96.20\%\), \(86.30\%\), \(92.65\%\), \(69.28\%\), \(68.04\%\) in terms of loss, accuracy, F1-score, precision, sensitivity, specificity, IoU, dice, hinge, and squared hinge. It is worth noting that the worst results were also obtained by the default UNet++ architecture.
The SpaSA meta-heuristic optimizer was utilized to optimize the pre-trained CNN models hyperparameters to achieve the learning, classification, and optimization phase. For the classification experiments, eight pre-trained CNN architectures are used, i.e., VGG16, VGG19, MobileNet, MobileNetV2, MobileNetV3Small, MobileNetV3Large, NasNetLarge, and NasNetMobile. For the “ISIC 2019 and 2020 Melanoma” dataset, the best reported overall accuracy from the applied CNN experiments is \(98.27\%\) by the MobileNet pre-trained model. The average accuracy was \(95.88\%\). The floored average TP, TN, FP, and FN were 11.250, 11.250, 198, and 198, respectively. Similarly, for the “Melanoma Classification (HAM10K)” dataset, the best reported overall accuracy from the applied CNN experiments is \(98.83\%\) by the MobileNet pre-trained model. The average accuracy was \(95.09\%\). The floored average TP, TN, FP, and FN were 9.867, 9.867, 117, and 117, respectively. For the “Skin diseases image” dataset, the best reported overall accuracy from the applied CNN experiments is \(85.87\%\) by the MobileNetV2 pre-trained model. The average accuracy was \(75.50\%\). The floored average TP, TN, FP, and FN were 22.130, 241.562, 2.518, and 4.990, respectively.
6.8 Related studies comparisons
Table 27 shows a comparison between the suggested approach and related studies concerning the same used datasets. It can be observed from the literature that there are different research works for skin cancer diagnosis. Additionally, using meta-heuristic algorithms for skin cancer diagnosis is increasing exponentially. In the current study, a hybrid algorithm using deep learning, transfer learning, and a recently proposed meta-heuristic algorithm named SpaSA has been suggested for performing skin cancer diagnosis. Many optimization approaches, including hybrid mechanisms, were previously used for skin cancer diagnosis; however, the SpaSA has not been utilized as far as we know concerning the published articles. Moreover, the suggested algorithm is trained and evaluated using 5 different datasets. Figure 12 shows a graphical comparison between the current study and the related studies.
6.9 Time complexity and other approaches remarks
The current study concentrated on skin cancer detection, classification, and segmentation. The classification was accomplished using 8 pre-trained CNNs, while the segmentation was done by employing 5 different U-Net models. The CNN hyperparameters optimization was performed using the SpaSA to gain the SOTA performance metrics. The target during the different experiments was to achieve high-performance metrics. The learning and processing time was high and hence was not declared exactly in this study. However, approximate times can be calculated. It is worth noting that the time depended mainly on the working environment. The current study worked on Google Colab. Assume that each CNN model takes one minute approximately. There are 15 hyperparameters to optimize using the SpaSA. The number of SpaSA iterations is set to 10, the SpaSA population size is set to 10, and the number of epochs is set to 5. Hence, there are \(10 \times 10 \times 5 = 500\) runs for each model to complete. The approximate time is 500 minutes (i.e., 8.33 hours) for a single model. We have 8 pre-trained models and 3 datasets. Hence, there are 24 experiments. The total approximate time can be 200 hours (i.e., 8.33 days). If the SpaSA is replaced with the grid search (GS) native search approach, the time would be more than that (e.g., months) as the GS searches for all possible combinations. From Table 11, we have 6 losses, 11 parameters optimizers, 60 dropouts, 12 batch sizes, 100 learning ratios, 4 scalers, 2 augmentation values, 45 rotations, 25 shifts in width, 25 shifts in height, 25 shears, 25 zooms, 2 horizontal flips, 2 vertical flips, and 15 brightness ranges. From that, we can obtain approximately 4E16 combinations which can last for more than 77 billion years.
7 Study limitations
Even though the proposed study demonstrated the potential of using deep learning models for detecting, classifying, and segmenting skin cancer, the suggested approach presents some limitations. The main limitation is the instantaneity, where the most time-consuming stage is the training of the classifier. The slow convergence of the boosting algorithm and high-dimensional features are the causes of this limitation. Another limitation includes that only 8 CNN architectures and 5 U-Net models are used.
8 Conclusions and future work
Automatic skin cancer detection and segmentation is an open-ended research area that steadily requires improvement. In this research, a methodological approach for classifying various skin images into their corresponding category with the help of convolution neural networks. CNN is a well-known SOTA approach for classifying images and big data. The study showed several works related to the current work. The deep learning-based skin cancer detection, classification, and segmentation system was developed. Different experiments were achieved and the results are reported. For the segmentation phase, concerning the “Skin cancer segmentation and classification” dataset, the best model was the U-Net++ with DenseNet201 as a backbone regarding the loss, accuracy, F1-score, AUC, IoU, and dice values. The achieved scores this architecture were \(94.16\%\), \(91.39\%\), \(99.03\%\), \(96.08\%\), \(96.41\%\), \(77.19\%\), \(75.47\%\) in terms of accuracy, F1-score, AUC, IoU, dice, hinge, and squared hinge. Additionally, concerning the “PH2” dataset, the best model was the Attention U-Net with DenseNet201 as a backbone regarding the loss, accuracy, F1, IoU, and dice values. The achieved scores by this architecture were 0.137, 94.75%, 92.65%, \(92.56\%\), \(92.74\%\), \(96.20\%\), \(86.30\%\), \(92.65\%\), \(69.28\%\), \(68.04\%\) in terms of loss, accuracy, F1-score, precision, sensitivity, specificity, IoU, dice, hinge, and squared hinge. The SpaSA meta-heuristic optimizer was utilized to optimize the pre-trained CNN models hyperparameters to achieve the learning, classification, and optimization phase. They were VGG16, VGG19, MobileNet, MobileNetV2, MobileNetV3Small, MobileNetV3Large, NasNetLarge, and NasNetMobile. The dataset was collected from different public sources. After collection, there are categorized into 2-class and 10-class datasets. For the “ISIC 2019 and 2020 Melanoma” dataset, the best reported overall accuracy from the applied CNN experiments is \(98.27\%\) by the MobileNet pre-trained model. Similarly, for the “Melanoma Classification (HAM10K)” dataset, the best reported overall accuracy from the applied CNN experiments is \(98.83\%\) by the MobileNet pre-trained model. For the “Skin diseases image” dataset, the best reported overall accuracy from the applied CNN experiments is \(85.87\%\) by the MobileNetV2 pre-trained model. The current study results were then compared with 13 prior related works. This showed that this study had outperformed numerous of the prior works. Even though there is a lot of work for the recognition and segmentation of skin cancer. There was a big challenge to get a suitable accuracy. Due to the lack of datasets in this area. In future studies, the plan is to (1) try other ML or DL techniques (2) work on enhancing the performance of the skin cancer segmentation phase, and (3) evaluate the system with further available datasets.
Data availability
The datasets, if existing, that are used, generated, or analyzed during the current study (A) if the datasets are owned by the authors, they are available from the corresponding author on reasonable request, (B) if the datasets are not owned by the authors, the supplementary information including the links and sizes is included in this published article.
References
Abdulazeem Y, Balaha HM, Bahgat WM, Badawy M (2021) Human action recognition based on transfer learning approach. IEEE Access 9:82058–82069
Adegun AA, Viriri S (2019) Deep learning-based system for automatic melanoma detection. IEEE Access 8:7160–7172
Ahmed N.J. (2021) Prescribing practices of medications in the outpatient dermatology department of a public hospital. J Pharm Res Int, pp 70–74
Ahsan MM, Mahmud M, Saha PK, Gupta KD, Siddique Z (2021) Effect of data scaling methods on machine learning algorithms and model performance. Technologies 9(3):52
Albahli S, Nida N, Irtaza A, Yousaf MH, Mahmood MT (2020) Melanoma lesion detection and segmentation using YOLOv4-darknet and active contour. IEEE Access 8:198403–198414
Alheejawi S, Berendt R, Jha N, Maity SP, Mandal M (2021) Detection of malignant melanoma in H &E-stained images using deep learning techniques. Tissue Cell 73:101659
Alsallakh B, Kokhlikyan N, Miglani V, Yuan J, Reblitz-Richardson O (2020) Mind the pad–cnns can develop blind spots. arXiv preprint arXiv:2010.02178
American Society of Clinical Oncology (2021). Skin Cancer (Non-Melanoma). http://www.cancer.net/cancer-types/skin-cancer-non-melanoma. Accessed: 24 Dec 2021
Arivuselvam B et al (2021) Skin cancer detection and classification using svm classifier. Turk J Comput Math Edu(TURCOMAT) 12(13):1863–1871
Astorino A, Fuduli A, Veltri P, Vocaturo E (2020) Melanoma detection by means of multiple instance learning. Interdiscip Sci Comput Life Sci 12(1):24–31
Badrinarayanan V, Kendall A, Cipolla R (2017) Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 39(12):2481–2495
Baghdadi NA, Malki A, Abdelaliem SF, Balaha HM, Badawy M, Elhosseini M (2022) An automated diagnosis and classification of COVID-19 from chest CT images using a transfer learning-based convolutional neural network. Comput Biol Med 144:105383
Bahgat WM, Balaha HM, AbdulAzeem Y, Badawy MM (2021) An optimized transfer learning-based approach for automatic diagnosis of COVID-19 from chest x-ray images. PeerJ Comput Sci 7:e555
Balaha HM, Ali HA, Badawy M (2021) Automatic recognition of handwritten Arabic characters: a comprehensive review. Neural Comput Appl 33(7):3011–3034
Balaha HM, Ali HA, Saraya M, Badawy M (2021) A new Arabic handwritten character recognition deep learning system (AHCR-DLS). Neural Comput Appl 33(11):6325–6367
Balaha HM, Ali HA, Youssef EK, Elsayed AE, Samak RA, Abdelhaleem MS, Tolba MM, Shehata MR, Mahmoud MR, Abdelhameed MM et al (2021) Recognizing Arabic handwritten characters using deep learning and genetic algorithms. Multimed Tools Appl 80(21):32473–32509
Balaha HM, Balaha MH, Ali HA (2021) Hybrid COVID-19 segmentation and recognition framework (HMB-HCF) using deep learning and genetic algorithms. Artif Intell Med 119:102156
Balaha HM, El-Gendy EM, Saafan MM (2021) Covh2sd: A COVID-19 detection approach based on Harris hawks optimization and stacked deep learning. Expert Syst Appl 186:115805
Balaha HM, El-Gendy EM, Saafan MM (2022) A complete framework for accurate recognition and prognosis of COVID-19 patients based on deep transfer learning and feature classification approach. Artif Intell Rev 55:1–46
Balaha HM, Saafan MM (2021) Automatic exam correction framework (AECF) for the MCQs, essays, and equations matching. IEEE Access 9:32368–32389
Balaha HM, Saif M, Tamer A, Abdelhay EH (2022) Hybrid deep learning and genetic algorithms approach (HMB-DLGAHA) for the early ultrasound diagnoses of breast cancer. Neural Comput Appl 34(11):8671–8695
Balaji V, Suganthi S, Rajadevi R, Kumar VK, Balaji BS, Pandiyan S (2020) Skin disease detection and segmentation using dynamic graph cut algorithm and classification through Naive Bayes classifier. Measurement 163:107922
Baskar R, Lee KA, Yeo R, Yeoh K-W (2012) Cancer and radiation therapy: current advances and future directions. Int J Med Sci 9(3):193
Binaghi E, Omodei M, Pedoia V, Balbi S, Lattanzi D, Monti E (2014) Automatic segmentation of MR brain tumor images using support vector machine in combination with graph cut. In: IJCCI (NCTA), pp 152–157
Binder M, Schwarz M, Winkler A, Steiner A, Kaider A, Wolff K, Pehamberger H (1995) Epiluminescence microscopy: a useful tool for the diagnosis of pigmented skin lesions for formally trained dermatologists. Arch Dermatol 131(3):286–291
Bronkhorst IH, Jager MJ (2013) Eye 27(2):217–223
Broomhead DS, Lowe D (1988) Radial basis functions, multi-variable functional interpolation and adaptive networks. Tech. rep, royal signals and radar establishment malvern (United Kingdom)
Brownlee J (2019) A gentle introduction to the rectified linear unit (ReLU). Mach Learn Mastery 6
Cancer.Net (2021) Melanoma: Statistics. https://www.cancer.net/cancer-types/melanoma/statistics. Accessed 24 Dec 2021
Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, Wang M (2021) Swin-unet: Unet-like pure transformer for medical image segmentation. arXiv preprint arXiv:2105.05537
Cao Z, Yang H, Zhao J, Guo S, Li L (2021) Attention fusion for one-stage multispectral pedestrian detection. Sensors 21(12):4184
Chan H-P, Lo S-CB, Sahiner B, Lam KL, Helvie MA (1995) Computer-aided detection of mammographic microcalcifications: pattern recognition with an artificial neural network. Med Phys 22(10):1555–1567
Chen L.-C, Papandreou G, Schroff F, Adam H (2017) Rethinking Atrous convolution for semantic image segmentation. arXiv preprint arXiv:1706.05587
Chollet F (2017) Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1251–1258
Ciregan D, Meier U, Schmidhuber J (2012) Multi-column deep neural networks for image classification. In: 2012 IEEE conference on computer vision and pattern recognition. IEEE, pp 3642–3649
Codella N, Rotemberg V, Tschandl P, Celebi M. E, Dusza S, Gutman D, Helba B, Kalloo A, Liopyris K, Marchetti M, et al. (2019) Skin lesion analysis toward melanoma detection 2018: a challenge hosted by the international skin imaging collaboration (isic). arXiv preprint arXiv:1902.03368
Codella NC, Gutman D, Celebi M.E, Helba B, Marchetti MA, Dusza SW, Kalloo A, Liopyris K, Mishra N, Kittler H, et al (2018) Skin lesion analysis toward melanoma detection: a challenge at the 2017 international symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018). IEEE, pp 168–172
Combalia M, Codella N.C, Rotemberg V, Helba B, Vilaplana V, Reiter O, Carrera C, Barreiro A, Halpern A.C., Puig S, et al. (2019) Bcn20000: Dermoscopic lesions in the wild. arXiv preprint arXiv:1908.02288
Coşkun M, Uçar A, Yildirim Ö, Demir Y (2017) Face recognition based on convolutional neural network. In: 2017 international conference on modern electrical and energy systems (MEES). IEEE, pp 376–379
Courbariaux M, Hubara I, Soudry D, El-Yaniv R, Bengio Y (2016) Binarized neural networks: Training deep neural networks with weights and activations constrained to+ 1 or-1. arXiv preprint arXiv:1602.02830
Danshina SD, Markov A, Huldani A (2020) Causes, symptoms, diagnosis and treatment of melanoma. Int J Pharm Res 12(3):903
Dauphin Y.N, De Vries H, Bengio Y (2015) Equilibrated adaptive learning rates for non-convex optimization. arXiv preprint arXiv:1502.04390
Davis LE, Shalin SC, Tackett AJ (2019) Current state of melanoma diagnosis and treatment. Cancer Biol Therapy 20(11):1366–1379
DeSantis CE, Miller KD, Dale W, Mohile SG, Cohen HJ, Leach CR, Sauer AG, Jemal A, Siegel RL (2019) Cancer statistics for adults aged 85 years and older. CA: A Cancer J Clin 69(6):452–467
DeVita VT, Lawrence TS, Rosenberg SA (2015) Cancer of the skin: cancer: principles & practice of oncology. Lippincott Williams & Wilkins, Philadelphia
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26(3):297–302
Dinnes J, di Ruffano LF, Takwoingi Y, Cheung ST, Nathan P, Matin RN, Chuchu N, Chan SA, Durack A, Bayliss SE et al (2019) Ultrasound, CT, MRI, or PET-CT for staging and re-staging of adults with cutaneous melanoma. Cochrane Database Syst Rev 7:CD012806
Esteva A, Kuprel B, Thrun S (2015) Deep networks for early stage skin disease and skin cancer classification. project report
Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, Abdel-Rahman O, Abdelalim A, Abdoli A, Abdollahpour I, Abdulle AS et al (2019) Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study. JAMA Oncol 5(12):1749–1768
Fujiyoshi H, Hirakawa T, Yamashita T (2019) Deep learning-based image recognition for autonomous driving. IATSS Res 43(4):244–252
Georgevici A.I, Terblanche M (2019) Neural networks and deep learning: a brief introduction
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y (2014) Generative adversarial nets. Adv Neural Inf Process Syst 27
Gu J, Wang Z, Kuen J, Ma L, Shahroudy A, Shuai B, Liu T, Wang X, Wang G, Cai J et al (2018) Recent advances in convolutional neural networks. Pattern Recogn 77:354–377
Gutman D, Codella N.C, Celebi E, Helba B, Marchetti M, Mishra N, Halpern A (2016) Skin lesion analysis toward melanoma detection: a challenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC). arXiv preprint arXiv:1605.01397
Hanley JA, McNeil BJ (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143(1):29–36
Hasan MK, Elahi MTE, Alam MA, Jawad MT, Martí R (2022) DermoExpert: Skin lesion classification using a hybrid convolutional neural network through segmentation, transfer learning, and augmentation. Inf Med Unlocked, p 100819
Hay R, Bendeck S. E, Chen S, Estrada R, Haddix A, McLeod T, Mahé A (2006) Skin diseases. Disease control priorities in developing countries. 2nd edn
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Hinton GE, Osindero S, Teh Y-W (2006) A fast learning algorithm for deep belief nets. Neural Comput 18(7):1527–1554
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9(8):1735–1780
Howard A, Sandler M, Chu G, Chen L.-C, Chen B, Tan M, Wang W, Zhu Y, Pang R, Vasudevan V, et al. (2019) Searching for mobilenetv3. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 1314–1324
Howard A.G, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, Andreetto M, Adam H (2017) Mobilenets: efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861
Huang G, Liu Z, Van Der Maaten L, Weinberger K Q (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Ibraheem N. A, Khan R.Z, Hasan M.M. (2013) Comparative study of skin color based segmentation techniques. Int J Appl Inf Syst, 5(10)
İlkin S, Gençtürk TH, Gülağız FK, Özcan H, Altuncu MA, Şahin S (2021) hybSVM: Bacterial colony optimization algorithm based SVM for malignant melanoma detection. Eng Sci Technol Int J 24:1059–1071
Jaccard P (1912) The distribution of the flora in the alpine zone. 1. New Phytol 11(2):37–50
Junayed M.S, Anjum N, Noman A, Islam B (2021) A deep CNN model for skin cancer detection and classification
Kaganami H.G, Beiji Z (2009) Region-based segmentation versus edge detection. In: 2009 fifth international conference on intelligent information hiding and multimedia signal processing. IEEE, pp 1217–1221
Kaur D, Kaur Y (2014) Various image segmentation techniques: a review. Int J Comput Sci Mob Comput 3(5):809–814
Kennedy C, Bajdik C, Willemze R, Bouwes Bavinck J (2005) Chemical exposures other than arsenic are probably not important risk factors for squamous cell carcinoma, basal cell carcinoma and malignant melanoma of the skin. Br J Dermatol 152(1):194–197
Khan MQ, Hussain A, Rehman SU, Khan U, Maqsood M, Mehmood K, Khan MA (2019) Classification of melanoma and nevus in digital images for diagnosis of skin cancer. IEEE Access 7:90132–90144
Kingma D. P, Ba J (2014) Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 25:1097–1105
Leachman SA, Carucci J, Kohlmann W, Banks KC, Asgari MM, Bergman W, Bianchi-Scarrà G, Brentnall T, Bressac-de Paillerets B, Bruno W et al (2009) Selection criteria for genetic assessment of patients with familial melanoma. J Am Acad Dermatol 61(4):677-e1
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444
Lee S, Kim H, Lieu QX, Lee J (2020) CNN-based image recognition for topology optimization. Knowl-Based Syst 198:105887
Lee TK (2001) Measuring border irregularity and shape of cutaneous melanocytic lesions. Ph.D. thesis, Citeseer
Li G, Jimenez G (2022) Optimal diagnosis of the skin cancer using a hybrid deep neural network and grasshopper optimization algorithm. Open Med 17(1):508–517
Loussaief S, Abdelkrim A (2018) Convolutional neural network hyper-parameters optimization based on genetic algorithms. Int J Adv Comput Sci Appl 9(10):252–266
Lu J, Behbood V, Hao P, Zuo H, Xue S, Zhang G (2015) Transfer learning using computational intelligence: a survey. Knowl-Based Syst 80:14–23
Madan V, Lear JT, Szeimies R-M (2010) Non-melanoma skin cancer. The Lancet 375(9715):673–685
Maniraj S, Maran PS (2022) A hybrid deep learning approach for skin cancer diagnosis using subband fusion of 3d wavelets. J Supercomput 78:1–16
Mar V, Roberts H, Wolfe R, English DR, Kelly JW (2013) Nodular melanoma: a distinct clinical entity and the largest contributor to melanoma deaths in Victoria, Australia. J Am Acad Dermatol 68(4):568–575
Markovic S.N, Erickson L.A, Rao R.D, McWilliams R.R, Kottschade L.A, Creagan E.T, Weenig R.H, Hand J.L., Pittelkow M.R, Pockaj B.A, et al. (2007) Malignant melanoma in the 21st century, part 1: epidemiology, risk factors, screening, prevention, and diagnosis. In: Mayo clinic proceedings, vol. 82, pp 364–380. Elsevier
McMahan H.B, Holt G, Sculley D, Young M, Ebner D, Grady J, Nie L, Phillips T, Davydov E, Golovin D, et al. (2013) Ad click prediction: a view from the trenches. In: Proceedings of the 19th ACM SIGKDD international conference on Knowledge discovery and data mining, pp 1222–1230
Mikołajczyk A, Grochowski M (2018) Data augmentation for improving deep learning in image classification problem. In: 2018 international interdisciplinary PhD workshop (IIPhDW), pp 117–122. IEEE
Milletari F, Navab N, Ahmadi S.-A (2016) V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: 2016 fourth international conference on 3D vision (3DV). IEEE, pp 565–571
Mohamad IB, Usman D (2013) Standardization and its effects on k-means clustering algorithm. Res J Appl Sci Eng Technol 6(17):3299–3303
Mukkamala M.C, Hein M(2017) Variants of rmsprop and adagrad with logarithmic regret bounds. In: International conference on machine learning. PMLR, pp. 2545–2553
Murugan A, Nair SAH, Kumar KS (2019) Detection of skin cancer using SVM, random forest and kNN classifiers. J Med Syst 43(8):1–9
Nabi J (2018) Machine learning-text processing. Towards Data Sci Retrieved 8(23):2019
Negin BP, Riedel E, Oliveria SA, Berwick M, Coit DG, Brady MS (2003) Symptoms and signs of primary melanoma: important indicators of Breslow depth. Cancer 98(2):344–348
Oktay O, Schlemper J, Folgoc L.L, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla N.Y, Kainz B, et al. (2018) Attention u-net: learning where to look for the pancreas. arXiv preprint arXiv:1804.03999
Pan SJ, Yang Q (2009) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359
Parkin D, Mesher D, Sasieni P (2011) cancers attributable to solar (ultraviolet) radiation exposure in the UK in 2010. British J Cancer 105(2):66–69
Pectasides D, Pectasides M, Economopoulos T (2006) Merkel cell cancer of the skin. Ann Oncol 17(10):1489–1495
Powers D.M (2020) Evaluation: from precision, recall and f-measure to roc, informedness, markedness and correlation. arXiv preprint arXiv:2010.16061
Pugazhenthi V, Naik S, Joshi A, Manerkar S, Nagvekar V, Naik K, Palekar C, Sagar K (2019) Skin disease detection and classification. Int J Adv Eng Res Sci 6(5):396–400
Ramachandran P, Zoph B, Le Q.V (2017) Searching for activation functions. arXiv preprint arXiv:1710.05941
Revathi V, Chithra A (2015) A review on segmentation techniques in skin lesion images. Int Res J Eng Technol 2(9):2598–2603
Rhodes AR (1995) Public education and cancer of the skin. what do people need to know about melanoma and nonmelanoma skin cancer? Cancer 75(S2):613–636
Rigel DS, Carucci JA (2000) Malignant melanoma prevention, early detection., and treatment in the 21st century. CA Cancer J Clin 50(4):215–236
Rogers HW, Weinstock MA, Feldman SR, Coldiron BM (2015) Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the us population, 2012. JAMA Dermatol 151(10):1081–1086
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241
Rotemberg V, Kurtansky N, Betz-Stablein B, Caffery L, Chousakos E, Codella N, Combalia M, Dusza S, Guitera P, Gutman D et al (2021) A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Scientific data 8(1):1–8
Ruder S (2016) An overview of gradient descent optimization algorithms. arXiv preprint arXiv:1609.04747
Rumelhart DE, Hinton GE, Williams RJ (1985) Learning internal representations by error propagation. California Univ San Diego La Jolla Inst for Cognitive Science, Tech. rep
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L.-C (2018) Mobilenetv2: Inverted residuals and linear bottlenecks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4510–4520
Sarkar D, Bali R, Ghosh T (2018) Hands-On Transfer Learning with Python: Implement advanced deep learning and neural network models using TensorFlow and Keras. Packt Publishing Ltd, Birmingham
Scherer D, Müller A, Behnke S (2010) Evaluation of pooling operations in convolutional architectures for object recognition. In: International conference on artificial neural networks. Springer, pp 92–101
Schuster M, Paliwal KK (1997) Bidirectional recurrent neural networks. IEEE Trans Signal Process 45(11):2673–2681
Seth R, Messersmith H, Kaur V, Kirkwood JM, Kudchadkar R, McQuade JL, Provenzano A, Swami U, Weber J, Alluri KC et al (2020) Systemic therapy for melanoma: Asco guideline. J Clin Oncol 38(33):3947–3970
Shan P, Wang Y, Fu C, Song W, Chen J (2020) Automatic skin lesion segmentation based on FC-DPN. Comput Biol Med 123:103762
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics, 2021. CA Cancer J Clin 71(1):7–33
Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A (2020) Colorectal cancer statistics, 2020. CA Cancer J Clin 70(3):145–164
Sikka K, Sinha N, Singh PK, Mishra AK (2009) A fully automated algorithm under modified fcm framework for improved brain mr image segmentation. Magn Reson Imaging 27(7):994–1004
Silveira M, Nascimento JC, Marques JS, Marçal AR, Mendonça T, Yamauchi S, Maeda J, Rozeira J (2009) Comparison of segmentation methods for melanoma diagnosis in dermoscopy images. IEEE J Sel Top Signal Process 3(1):35–45
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Singh KK, Singh A (2010) A study of image segmentation algorithms for different types of images. Int J Comput Sci Issues 7(5):414
Taha AA, Hanbury A (2015) Metrics for evaluating 3d medical image segmentation: analysis, selection, and tool. BMC Med Imaging 15(1):1–28
Takarli F, Aghagolzadeh A, Seyedarabi H (2016) Combination of high-level features with low-level features for detection of pedestrian. Signal Image Video Process 10(1):93–101
Tariman J.D, Berry D.L, Cochrane B, Doorenbos A, Schepp K (2012) Physician, patient and contextual factors affecting treatment decisions in older adults with cancer: a literature review. In: Oncology nursing forum, vol 39 (p E70). NIH Public Access
Tato A, Nkambou R (2018) Improving Adam optimizer
Theodoridis S (2015) Neural networks and deep learning. Mach Learn, pp 875–936
Thomas L, Tranchand P, Berard F, Secchi T, Colin C, Moulin G (1998) Semiological value of ABCDE criteria in the diagnosis of cutaneous pigmented tumors. Dermatology 197(1):11–17
Torrey L, Shavlik J (2010) Transfer learning. In: Handbook of research on machine learning applications and trends: algorithms, methods, and techniques. IGI global, pp 242–264
Tschandl P, Rosendahl C, Kittler H (2018) The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data 5:180161
Vani R, Kavitha J, Subitha D (2021) Novel approach for melanoma detection through iterative deep vector network. J Ambient Intell Hum Comput, pp 1–10
Verma S (2018) Understanding different loss functions for neural networks. Towards Data Sci
Wang Y-H (2010) Tutorial: image segmentation. National Taiwan University, Taipei, pp 1–36
Ward R, Wu X, Bottou L (2019) Adagrad stepsizes: Sharp convergence over nonconvex landscapes. In: International conference on machine learning. PMLR, pp 6677–6686
Weinberg RA (1996) How cancer arises. Sci Am 275(3):62–70
Wysocki AB (1999) Skin anatomy, physiology, and pathophysiology. Nurs Clin North Am 34(4):777–97
Xiang T, Wang J, Liao X (2007) An improved particle swarm optimizer with momentum. In: 2007 IEEE congress on evolutionary computation. IEEE, pp 3341–3345
Xue J, Shen B (2020) A novel swarm intelligence optimization approach: sparrow search algorithm. Syst Sci Control Eng 8(1):22–34
Zeiler M.D (2012) Adadelta: an adaptive learning rate method. arXiv preprint arXiv:1212.5701
Zhang Y, Wallace B (2015) A sensitivity analysis of (and practitioners’ guide to) convolutional neural networks for sentence classification. arXiv preprint arXiv:1510.03820
Zhang YJ (1996) A survey on evaluation methods for image segmentation. Pattern Recogn 29(8):1335–1346
Zhao W, Zhang Z, Wang L (2020) Manta ray foraging optimization: an effective bio-inspired optimizer for engineering applications. Eng Appl Artif Intell 87:103300
Zhou Z, Siddiquee M.M.R, Tajbakhsh N, Liang J (2018) Unet++: A nested u-net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support, pp 3–11. Springer
Zoph B, Vasudevan V, Shlens J, Le Q.V (2018) Learning transferable architectures for scalable image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 8697–8710
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
All the authors have participated in writing the manuscript and have revised the final version. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest exists. We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Intellectual property
We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, concerning intellectual property. In so doing, we confirm that we have followed the regulations of our institutions concerning intellectual property.
Research ethics
We further confirm, if existing, that any aspect of the work covered in this manuscript that has involved human patients has been conducted with the ethical approval of all relevant bodies and that such approvals are acknowledged within the manuscript. Written consent to publish potentially identifying information, such as details of the case and photographs, was obtained from the patient(s) or their legal guardian(s).
Authorship
We confirm that the manuscript has been read and approved by all named authors. We confirm that the order of authors listed in the manuscript has been approved by all named authors.
Human participants and/or animals
The current study does not contain any studies with human participants and/or animals performed by any of the authors.
Consent to participate
There is no informed consent for the current study.
Consent for publication
Not applicable.
Contact with the editorial office
The “Corresponding Author” who is declared on the title page. This author submitted this manuscript using his account in the editorial submission system. (A) We understand that the “Corresponding Author” is the sole contact for the Editorial process (including the editorial submission system and direct communications with the office). He is responsible for communicating with the other authors about progress, submissions of revisions, and final approval of proofs. (B) We confirm that the email address shown below is accessible by the “Corresponding Author” and is the address to which “Corresponding Author’s” editorial submission system account is linked and has been configured to accept email from the editorial office (email: hossam.m.balaha@mans.edu.eg).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendices
1.1 Table of Abbreviations
Table 28 shows the abbreviations and the corresponding meaning. They are sorted alphabetically in ascending order.
1.2 Author agreement statement
We, the undersigned authors, declare that this manuscript is original, has not been published before, and is not currently being considered for publication elsewhere. We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us. We understand that the “corresponding author” is the sole contact for the editorial process. He is responsible for communicating with the other authors about progress, submissions of revisions, and final approval of proofs.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Balaha, H.M., Hassan, A.ES. Skin cancer diagnosis based on deep transfer learning and sparrow search algorithm. Neural Comput & Applic 35, 815–853 (2023). https://doi.org/10.1007/s00521-022-07762-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07762-9