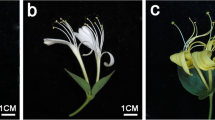
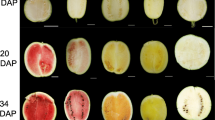
Abstract
Key message
α-Carotene and β-carotene were the major substances of petal coloration in Osmanthus fragrans ‘Yanhong Gui’. We identified 15 key carotenogenic genes involved in petal coloration of O. fragrans ‘Yanhong Gui’.
Abstract
Osmanthus fragrans Lour. is a famous traditional woody flower in China. Carotenoids are the major coloring factor in the petals of O. fragrans. However, little information is available on the transcriptional regulation of carotenogenic genes on petal coloration of orange-red-flowered cultivar. In this study, petal color, carotenoid content, transcriptome dynamics of flower buds and expression of key carotenogenic genes were analyzed by use of an orange-red-flowered cultivar ‘Yanhong Gui’ of O. fragrans. Results showed that α-carotene and β-carotene were identified as the major substances of petal coloration in O. fragrans ‘Yanhong Gui’. Using transcriptome sequencing technology, about 54.79, 57.79, 54.71 and 58.04 million raw reads were obtained and assembled into 175,320, 177,162, 175,248 and 161,902 unigenes from four cDNA libraries, respectively. Out of these assembled unigenes, a total of 129,670, 65,655 and 49,837 unigenes were assigned to non-redundant protein, Gene Ontology and Cluster of Orthologous Groups of proteins database, respectively. Furthermore, out of 46 unigenes which were identified to be carotenoid-related genes, 35 differentially expressed genes (DEGs) were generated with the threshold of fold change ≥2 or ≤0.5. Based on the expression levels, 35 DEGs were grouped into five clusters using the hierarchical method. Taken petal color development and carotenoid content into consideration, we identified 27 candidate key DEGs involved in carotenoid metabolism during flower bud development. Further analysis with quantitative real-time PCR generated 15 key carotenogenic genes involved in petal coloration of O. fragrans ‘Yanhong Gui’. In addition to providing valuable sequence resource of O. fragrans, all the results and analysis are valuable resources for better understanding of carotenoids metabolism in this plant.










Similar content being viewed by others
References
Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP (2002) Gene expression during the life cycle of Drosophila melanogaster. Science 297:2270–2275
Baldermann S, Kato M, Kurosawa M, Kurobayashi Y, Fujita A, Fleischmann P, Watanabe N (2010) Functional characterization of a carotenoid cleavage dioxygenase 1 and its relation to the carotenoid accumulation and volatile emission during the floral development of Osmanthus fragrans Lour. J Exp Bot 61:2967–2977
Britton G, Liaaen-Jensen S, Pfander H (eds) (2004) Carotenoids handbook. Birkhäuser, Basel
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116
Chappell J (1995) Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Annu Rev Plant Physiol Plant Mol Biol 46:521–547
Chen H, Zha J, Liang X, Bu J, Wang M, Wang Z (2013) Sequencing and de novo assembly of the Asian clam (Corbicula fluminea) transcriptome using the Illumina GAIIx method. PLoS One 8:e79516
Chen Y, Mao Y, Liu H, Yu F, Li S, Yin T (2014) Transcriptome analysis of differentially expressed genes relevant to variegation in peach flowers. PLoS One 9:e90842
Clotault J, Peltier D, Berruyer R, Thomas M, Briard M, Geoffriau E (2008) Expression of carotenoid biosynthesis genes during carrot root development. J Exp Bot 59:3563–3573
Consortium GO (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32:D258–D261
Del Villar-Martínez AA, García-Saucedo PA, Carabez-Trejo A, Cruz-Hernández A, Paredes-López O (2005) Carotenogenic gene expression and ultrastructural changes during development in marigold. J Plant Physiol 162:1046–1056
Delpino-Rius A, Eras J, Marsol-Vall A, Vilaró F, Balcells M, Canela-Garayoa R (2014) Ultra performance liquid chromatography analysis to study the changes in the carotenoid profile of commercial monovarietal fruit juices. J Chromatogr A 1331:90–99
Eisenreich W, Rohdich F, Bacher A (2001) Deoxyxylulose phosphate pathway to terpenoids. Trends Plant Sci 6:78–84
Gao J, Zhang Y, Zhang C, Qi F, Li X, Mu S, Peng Z (2014) Characterization of the floral transcriptome of Moso bamboo (Phyllostachys edulis) at different flowering developmental stages by transcriptome sequencing and RNA-seq analysis. PLoS One 9:e98910
Green B, Durnford D (1996) The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 47:685–714
Han Y, Li L, Dong M, Yuan W, Shang F (2013) cDNA cloning of the phytoene synthase (PSY) and expression analysis of PSY and carotenoid cleavage dioxygenase genes in Osmanthus fragrans. Biologia 68:258–263
Han Y, Wang X, Chen W, Dong M, Yuan W, Liu X, Shang F (2014) Differential expression of carotenoid-related genes determines diversified carotenoid coloration in flower petal of Osmanthus fragrans. Tree Genet Genomes 10:329–338
Han Y, Chen W, Yang F, Wang X, Dong M, Zhou P, Shang F (2015) cDNA-AFLP analysis on 2 Osmanthus fragrans cultivars with different flower color and molecular characteristics of OfMYB1 gene. Trees Struct Funct 29:931–940
Huang F-C, Horváth G, Molnár P, Turcsi E, Deli J, Schrader J, Sandmann G, Schmidt H, Schwab W (2009) Substrate promiscuity of RdCCD1, a carotenoid cleavage oxygenase from Rosa damascena. Phytochemistry 70:457–464
Jing G, Li T, Qu H, Yun Z, Jia Y, Zheng X, Jiang Y (2015) Carotenoids and volatile profiles of yellow- and red-fleshed papaya fruit in relation to the expression of carotenoid cleavage dioxygenase genes. Postharvest Biol Technol 109:114–119
Kishimoto S, Ohmiya A (2009) Studies on carotenoids in the petals of Compositae plants. J Japan Soc Hortic Sci 78:263–272
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Method Enzymol 148:350–382
Lichtenthaler HK (1999) The 1-deoxy-d-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu Rev Plant Biol 50:47–65
Liu D, Sui S, Ma J, Li Z, Guo Y, Luo D, Yang J, Li M (2014) Transcriptomic analysis of flower development in wintersweet (Chimonanthus praecox). PLoS One 9:e86976
Lomax J (2005) Get ready to GO! A biologist’s guide to the Gene Ontology. Brief Bioinform 6:298–304
Ma J, Li J, Zhao J, Zhou H, Ren F, Wang L, Gu C, Liao L, Han Y (2014) Inactivation of a gene encoding carotenoid cleavage dioxygenase (CCD4) leads to carotenoid-based yellow coloration of fruit flesh and leaf midvein in peach. Plant Mol Biol Rep 32:246–257
Moehs CP, Li T, Osteryoung KW, Dellapenna D (2001) Analysis of carotenoid biosynthetic gene expression during marigold petal development. Plant Mol Biol 45:281–293
Mu HN, Li HG, Wang LG, Yang XL, Sun TZ, Xu C (2014) Transcriptome sequencing and analysis of sweet osmanthus (Osmanthus fragrans Lour.). Genes Genomics 36:777–788
Nakkanong K, Yang JH, Zhang MF (2012) Carotenoid accumulation and carotenogenic gene expression during fruit development in novel interspecific inbred squash lines and their parents. J Agric Food Chem 60:5936–5944
Niyogi KK (2000) Safety valves for photosynthesis. Curr Opin Plant Biol 3:455–460
Novaes E, Drost DR, Farmerie WG, Pappas GJ, Grattapaglia D, Sederoff RR, Kirst M (2008) High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genom 9:312
Ohmiya A (2013) Qualitative and quantitative control of carotenoid accumulation in flower petals. Sci Hortic Amsterdam 163:10–19
Romualdi C, Bortoluzzi S, d’Alessi F, Danieli GA (2003) IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics 12:159–162
Ruiz-Sola MÁ, Rodríguez-Concepción M (2012) Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book 10:e0158
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 36:1101–1108
van Breemen RB, Dong L, Pajkovic ND (2012) Atmospheric pressure chemical ionization tandem mass spectrometry of carotenoids. Int J Mass Spectrom 312:163–172
Vêncio RZ, Brentani H, Pereira CA (2003) Using credibility intervals instead of hypothesis tests in SAGE analysis. Bioinformatics 19:2461–2464
Walter MH, Strack D (2011) Carotenoids and their cleavage products: biosynthesis and functions. Nat Prod Rep 28:663–692
Xiang Q, Liu Y (eds) (2007) An illustrated monograph of the sweet osmanthus variety in China. Zhejiang Science and Technology Press, Hangzhou
Xie F, Burklew CE, Yang Y, Liu M, Xiao P, Zhang B, Qiu D (2012) De novo sequencing and a comprehensive analysis of purple sweet potato (Impomoea batatas L.) transcriptome. Planta 236:101–113
Xu H, Gao Y, Wang J (2012) Transcriptomic analysis of rice (Oryza sativa) developing embryos using the RNA-Seq technique. PLoS One 7:e30646
Yamamizo C, Kishimoto S, Ohmiya A (2010) Carotenoid composition and carotenogenic gene expression during Ipomoea petal development. J Exp Bot 61:709–719
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293–W297
Young PR, Lashbrooke JG, Alexandersson E, Jacobson D, Moser C, Velasco R, Vivier MA (2012) The genes and enzymes of the carotenoid metabolic pathway in Vitis vinifera L. BMC Genom 13:243
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829
Zhang C, Wang Y, Fu J, Dong L, Gao S, Du D (2014) Transcriptomic analysis of cut tree peony with glucose supply using the RNA-Seq technique. Plant Cell Rep 33:111–129
Zhao D, Jiang Y, Ning C, Meng J, Lin S, Ding W, Tao J (2014) Transcriptome sequencing of a chimaera reveals coordinated expression of anthocyanin biosynthetic genes mediating yellow formation in herbaceous peony (Paeonia lactiflora Pall.). BMC Genom 15:689
Zhou H, Cheng F-Y, Wang R, Zhong Y, He C (2013) Transcriptome comparison reveals key candidate genes responsible for the unusual reblooming trait in tree peonies. PLoS One 8:e79996
Zhu C, Yamamura S, Koiwa H, Nishihara M, Sandmann G (2002) cDNA cloning and expression of carotenogenic genes during flower development in Gentiana lutea. Plant Mol Biol 48:277–285
Zhu C, Yamamura S, Nishihara M, Koiwa H, Sandmann G (2003) cDNAs for the synthesis of cyclic carotenoids in petals of Gentiana lutea and their regulation during flower development. Biochim Biophys Acta 1625:305–308
Zhu C, Bai C, Sanahuja G, Yuan D, Farré G, Naqvi S, Shi L, Capell T, Christou P (2010) The regulation of carotenoid pigmentation in flowers. Arch Biochem Biophys 504:132–141
Zhu H, Chen M, Wen Q, Li Y (2015) Isolation and characterization of the carotenoid biosynthetic genes LCYB, LCYE and CHXB from strawberry and their relation to carotenoid accumulation. Sci Hortic Amsterdam 182:134–144
Acknowledgments
This research was supported by the National Natural Science Foundation of China (Grant No. 31501790, 31170656 and 31101571), Zhejiang Provincial Natural Science Foundation of China (Grant No. LQ15C160004 and LQ16C160003), Zhejiang Provincial Major Program of New Cultivar Breeding (Grant No. 2012C12909-9 and 2012C12909-19) and Open Foundation of Top Key Discipline of Forestry, Zhejiang Province (KF201321).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by J.E. Carlson.
Electronic supplementary material
Below is the link to the electronic supplementary material.
468_2016_1359_MOESM1_ESM.jpg
Supplementary Fig. 1 HPLC separation of carotenoids of extracts of O. fragrans ‘Yanhong Gui’ petals at different developmental stage. Peak 7 (P7) represents the peak of internal standard β-apo-8′-carotenal, and other peaks represent different carotenoid compositions. S1, the outer bud scales unfurled and the inner bud scales still furled; S2, the inner bud scales unfurled and the bud remained spindle-shaped; S3, the bud became globular-shaped and the inside bracts covering the inflorescence was visible; S4, the inflorescence burst through bracts and the florets closely crowded; S5, initial flowering stage; S6, full flowering stage (JPEG 431 kb)
468_2016_1359_MOESM2_ESM.jpg
Supplementary Fig. 2 Twenty-one kinds of gene groups classified by their expression pattern during bud development. Groups are as follows: 1, up-invariable-up expression pattern; 2, up-invariable-invariable expression pattern; 3, up-invariable-down expression pattern; 4, up-down-up expression pattern; 5, up-down-invariable expression pattern; 6, up-down-down expression pattern; 7, invariable-up-up expression pattern; 8, invariable-up-invariable expression pattern; 9, invariable-up-down expression pattern; 10, invariable-invariable-up expression pattern; 11, invariable-invariable-invariable expression pattern; 12, invariable-invariable-down expression pattern; 13, invariable-down-up expression pattern; 14, invariable-down-invariable expression pattern; 15, invariable-down-down expression pattern; 16, down-up-up expression pattern; 17, down-up-invariable expression pattern; 18, down-up-down expression pattern; 19, down-invariable-up expression pattern; 20, down-invariable-invariable expression pattern; 21, down-invariable-down expression pattern (JPEG 5378 kb)
Rights and permissions
About this article
Cite this article
Zhang, C., Wang, Y., Fu, J. et al. Transcriptomic analysis and carotenogenic gene expression related to petal coloration in Osmanthus fragrans ‘Yanhong Gui’. Trees 30, 1207–1223 (2016). https://doi.org/10.1007/s00468-016-1359-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-016-1359-8