Abstract
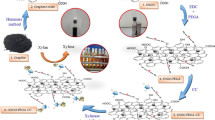
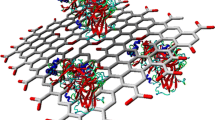
Herein, we propose the synthesis and characterization of graphene for the immobilization of β-galactosidase for improved galacto-oligosaccharide (GOS) production. The size of synthesized graphene was observed to be 25 nm by TEM analysis while interaction of enzyme with the nanosupport was observed by FTIR spectroscopy. Docking was obtained using molecular docking program Dock v.6.5 while the visual analyses and illustration of protein–ligand complex were investigated by utilizing chimera v.1.6.2 and PyMOL v.1.3 softwares. Immobilized β-galactosidase (IβG) showed improved stability against various physical and chemical denaturants. K m of IβG was increased to 6.41 mM as compared to 2.38 mM of soluble enzyme without bringing significant change in V max value. Maximum GOS content also registered an increase in lactose conversion. The maximum GOS production was achieved by immobilized enzyme at specific temperature and time. Hence, the developed nanosupport can be further exploited for developing a biosensor involving β-galactosidase or for immobilization of other industrially/therapeutically important enzymes.








Similar content being viewed by others
Abbreviations
- GO:
-
Graphene oxide
- GOS:
-
Galacto-oligosaccharides
- IβG:
-
Immobilized β-galactosidase
- SβG:
-
Soluble β-galactosidase
References
Kiehl K, Straube T, Opwis K, Gutmann JS (2015) Strategies for permanent immobilization of enzymes on textile carriers. Eng Life Sci 15:622–626
Mohamad NR, Marzuki NHC, Buang NA, Huyop F, Wahab RA (2015) An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol Biotechnol Equip 29:205–220
Homaei AA, Sariri R, Vianello F, Stevanato R (2013) Enzyme immobilization: an update. J Chem Biol 6:185–205
Ansari SA, Husain Q (2012) Potential application of enzymes immobilized on/in nano materials: a review. Biotechnol Adv 30:512–523
Cipolatti EP, Silva MJA, Klein MP, Federn V, Feltes MMC, Oliveira JV (2014) Current status and trends in enzymatic nanoimmobilization. J Mol Catal B Enzym 99:56–67
Fei L, Perrett S (2009) Effect of nanoparticles on protein folding and fibrillogenesis. Int J Mol Sci 10:646–655
Goenka S, Sant V, Sant S (2014) Graphene-based nanomaterials for drug delivery and tissue engineering. J Control Release 173:75–88
Soldano C, Mahmood A, Dujardin E (2010) Production, properties and potential of graphene. Carbon 48:2127–2150
Singh V, Joung D, Zhai L, Das S, Khondaker SI, Seal S (2011) Graphene based materials: past, present and future. Prog Mat Sci 56:1178–1271
Georgakilas V, Otyepka M, Bourlinos AB, Chandra V, Kim N, Kemp KC, Hobza P, Zboril R, Kim KS (2012) Functionalization of graphene: covalent and non-covalent approaches, derivatives and application. Chem Rev 112:6156–6214
Hauser AS, Windshugel B (2016) A benchmark data set for assessment of peptide docking performance. J Chem Inf Mod. doi:10.1021/acs.jcim.5b00234
Panneerselvam S, Choi S (2014) Nanoinformatics: emerging databases and available tools. Int J Mol Sci 15:7158–7182
Suresh PS, Kumar A, Kumar R, Singh VP (2008) An insilico approach to bioremediation: laccase as a case study. J Mol Graph Mod 26:845–849
Morris GM, Huey R, Olson AJ (2008) Using autoDock for ligand-receptor docking. Curr Protoc Bioinform 8(8):14
Yang ST, Wang H, Guo L, Gao Y, Liu Y, Cao A (2008) Interaction of fullerenol with lysozyme investigated by experimental and computational approaches. Nanotechnology 19:395101
Diez-Municio M, Herrero M, Olano A, Moreno FJ (2014) Synthesis of novel bioactive lactose-derived oligosaccharides by microbial glycoside hydrolases. Microb Biotechnol 7:315–331
Otieno DO (2010) Synthesis of β-galactooligosaccharides from lactose using microbial β-galactosidases. Comp Rev Food Sci Food Saf 9:471–482
Zhao X, Liu L, Li X, Zeng J, Jia X, Liu P (2014) Biocompatible graphene oxide nanoparticle-based drug delivery platform for tumor microenvironment-responsive triggered release of doxorubicin. Langmuir 30:10419–10429
Ewing TJ, Makino S, Skillman AG, Kuntz ID (2001) DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aid Mol Des 15:411–428
DeLano WL (2002) The PyMOL molecular graphics system. DeLano Scientific, San Carlos
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8:127–134
Laskowski RA, Swindells MB (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786
Ansari SA, Satar R, Zaidi SK (2015) Carboxylation of silver nanoparticles for the immobilization of β-galactosidase and its efficacy in galacto-oligosaccharides production. Quim Nova 38:387–392
Fai AEC, Postore GM (2015) Galactooligosaccharides: production, health benefits, application to foods and perspectives. Agropecuaria 6:69–81
Neri DFM, Balcao VM, Costa RS, Rocha ICAP, Ferreria EMFC, Torres DPM, Rodriguez LRM, Carvalho LB Jr, Teixeria JA (2009) Galacto-oligosaccharides production during lactose hydrolysis by free Aspergillus oryzae β-galactosidase and immobilized on magnetic polysiloxane-polyvinyl alcohol. Food Chem 115:92–99
Srivastava G, Singh K, Talat M, Srivastava ON, Kayastha AM (2014) Functionalized graphene sheets as immobilization matrix for fenugreek β-amylase: enzyme kinetics and stability studies. PLoS ONE 9:e113408
Hsu CA, Lee SL, Chou CC (2007) Enzymatic production of galactooligosaccharides by beta-galactosidase from Bifidobacterium longum BCRC 15708. J Agric Food Chem 21:2225–2230
Barbara RC, Miguel AA, Lucia FA, Roseri B, Ana P, Jesus JB, Dietmar H, Antonio OO, Maria FL, Francisco JP (2011) Production of galacto-oligosaccharides by β-galactosidase from Kluyveromyces lactis: comparative analysis of permeabilized cells versus soluble enzyme. J Agric Food Chem 59:10477–10484
Warmerdam A, Boom RM, Janssen AEM (2013) β galactosidase stability at high substrate concentrations. Springer Plus 2:402–409
Palai T, Kumar A, Bhattacharya BK (2015) Kinetic studies and model development for the formation of galacto-oligosaccharides from lactose using synthesized thermo-responsive bioconjugates. Enzym Microb Technol 70:42–49
Albayrak N, Yang ST (2002) Production of galacto-oligosaccharides from lactose by Aspergillus oryzae beta-galactosidase immobilized on cotton cloth. Biotechnol Bioeng 5:8–19
Pruksasri S, Nguyen TH, Haltrich D, Novalin S (2015) Fractionation of a galacto-oligosaccharides solution at low and high temperature using nanofiltration. Sep Purif Technol 4:124–130
Sen P, Bhattacharjee C, Bhattacharya P (2016) Experimental studies and two-dimensional modeling of a packed bed bioreactor used for production of galacto-oligosaccharides (GOS) from milk whey. Bioprocess Biosyst Eng. doi:10.1007/s00449-015-1516-2
Acknowledgments
The authors are thankful to Prof. Waseem Ahmad (Center of Excellence in Genomic and Medicine Research, King Abdulaziz University, Saudi Arabia) for going through the manuscript critically and suggesting relevant changes.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Satar, R., Ismail, S.A., Rehan, M. et al. Elucidating the binding efficacy of β-galactosidase on graphene by docking approach and its potential application in galacto-oligosaccharide production. Bioprocess Biosyst Eng 39, 807–814 (2016). https://doi.org/10.1007/s00449-016-1560-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-016-1560-6