Abstract
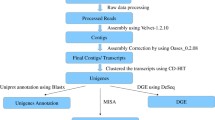
Radish is an important root vegetable crop with high nutritional, economic, and medicinal value. Lignin is an important secondary metabolite possessing a great effect on plant growth and product quality. To date, lignin biosynthesis-related genes have been identified in some important plant species. However, little information on characterization of critical genes involved in plant lignin biosynthesis is available in radish. In this study, a total of 71,148 transcripts sequences were obtained from radish root, of which 66 assembled unigenes and ten candidate genes were identified to be involved in lignin monolignol biosynthesis. Full-length cDNA sequences of seven randomly selected genes were isolated and sequenced from radish root, and the assembled unigenes covered more than 80% of their corresponding cDNA sequences. Moreover, the lignin content gradually accumulated in leaf during the developmental stages, and it increased from pre-cortex to cortex splitting stage, followed by a decrease at thickening stage and then increased at mature stage in root. RT-qPCR analysis revealed that all these genes except RsF5H exhibited relatively low expression level in root at thickening stage. The expression profiles of Rs4CL5, RsCCoAOMT1, and RsCOMT genes were consistent with the changes of root lignin content, implying that these candidate genes may play important roles in lignin formation in radish root. These findings would provide valuable information for identification of lignin biosynthesis-related genes and facilitate dissection of molecular mechanism underlying lignin biosynthesis in radish and other root vegetable crops.





Similar content being viewed by others
References
Abdulrazzak N, Pollet B, Ehlting J, Larsen K, Asnaghi C, Ronseau S, Proux C, Erhardt M, Seltzer V, Renou J, Ullmann P, Pauly M, Lapierre C, Werck-Reichhart D (2006) A coumaroyl -ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol 140:30–48
Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19:148–162
Boudet AM (2000) Lignins and lignification: selected issues. Plant Physiol Biochem 38:81–96
Coleman HD, Park JY, Nair R, Chapple C, Mansfield SD (2008) RNAi-mediated suppression of p-coumaroyl-CoA 3′-hydroxylase in hybrid poplar impacts lignin deposition and soluble secondary metabolism. Proc Natl Acad Sci USA 105:4501–4506
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Dhingra D, Michael M, Rajput H, Patil RT (2012) Dietary fibre in foods: a review. J Food Sci Technol 49:255–266
Dixon RA, Chen F, Guo D, Parvathi K (2001) The biosynthesis of monolignols: a “metabolic grid”, or independent pathways to guaiacyl and syringyl units? Phytochemistry 57:1069–1084
Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, Cullis IF, Zhuang J, Kaneda M, Mansfield SD, Samuels L, Ritland K, Ellis BE, Bohlmann J, Douglas CJ (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J 42:618–640
Fagerstedt KV, Saranpää P, Tapanila T, Immanen J, Serra JAA, Nieminen K (2015) Determining the composition of lignins in different tissues of silver birch. Plants 4:183–195
Franke R, McMichael CM, Meyer K, Shirley AM, Cusumano JC, Chapple C (2000) Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J 22:223–234
Franke R, Hemm MR, Denault JW, Ruegger MO, Humphreys JM, Chapple C (2002) Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J 30:47–59
Gosselink RJA, De Jong E, Guran B, Abächerli A (2004) Co-ordination network for lignin—standardisation, production and applications adapted to market requirements (EUROLIGNIN). Ind Crop Prod 20:121–129
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson D, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Gutiérrez RM, Perez RL (2004) Raphanus sativus (Radish): their chemistry and biology. Sci World J 4:811–837
Hahn DA, Ragland GJ, Shoemaker DD, Denlinger DL (2009) Gene discovery using massively parallel pyrosequencing to develop ESTs for the flesh fly Sarcophaga crassipalpis. BMC Genom 10:234
Harrington MJ, Mutwil M, Barrière Y, Sibout R (2012) Molecular biology of lignification in grasses. Adv Bot Res 61:77–112
Huang HH, Xu LL, Tong ZK, Lin EP, Liu QP, Cheng LJ, Zhu MY (2012) De novo characterization of the Chinese fir (Cunninghamia lanceolata) transcriptome and analysis of candidate genes involved in cellulose and lignin biosynthesis. BMC Genom 13:648
Hyun TK, Rim Y, Jang HJ, Kim CH, Park J, Kumar R, Lee S, Kim BC, Bhak J, Nguyen-Quoc B, Kim SW, Lee SY, Kim JY (2012) De novo transcriptome sequencing of Momordica cochinchinensis to identify genes involved in the carotenoid biosynthesis. Plant Mol Biol 79:413–427
Jia XL, Wang GL, Xiong F, Yu XR, Xu ZS, Wang F, Xiong AS (2015) De novo assembly, transcriptome characterization, lignin accumulation, and anatomic characteristics: novel insights into lignin biosynthesis during celery leaf development. Sci Rep 5:8259
Kim JW, Kim MB, Lim SB (2015) Formation and stabilization of raphasatin and sulforaphene from radish roots by endogenous enzymolysis. Prev Nutr Food Sci 20:119–125
Kitashiba H, Li F, Hirakawa H, Kawanabe T, Zou Z, Hasegawa Y, Tonosaki K, Shirasawa S, Fukushima A, Yokoi S (2014) Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res 21:481–490
Koutaniemi S, Warinowski T, Kärkönen A, Alatalo E, Fossdal CG, Saranpää P, Laakso T, Fagerstedt KV, Simola LK, Paulin L, Rudd S, Teeri TH (2007) Expression profiling of the lignin biosynthetic pathway in Norway spruce using EST sequencing and real-time RT-PCR. Plant Mol Biol 65:311–328
Li D, Deng Z, Qin B, Liu X, Men Z (2012) De novo assembly and characterization of bark transcriptome using Illumina sequencing and development of EST-SSR markers in rubber tree (Hevea brasiliensis Muell. Arg.). BMC Genom 13:192
Litvak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Mitjans M, Vinardell MP (2005) Biological activity and health benefits of lignans and lignins. Trends Comp Biochem Physiol 11:55–62
Montoya-Burgos JI, Foulon A, Bahechar I (2010) Transcriptome screen for fast evolving genes by Inter-Specific Selective Hybridization (ISSH). BMC Genom 11:126
Moura JCMS, Bonine CAV, De Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P (2010) Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol 52:360–376
Nguyen TN, Son SH, Jordan MC, Levin DB, Ayele BT (2016) Lignin biosynthesis in wheat (Triticum aestivum L.): its response to waterlogging and association with hormonal levels. BMC Plant Biol 16:28
Novaes E, Kirst M, Chiang V, Winter-Sederoff H, Sederoff R (2010) Lignin and biomass: a negative correlation for wood formation and lignin content in trees. Plant Physiol 154:555–561
Olsen KM, Lea US, Slimestad R, Verheul M, Lillo C (2008) Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J Plant Physiol 165:1491–1499
Reddy MS, Chen F, Shadle G, Jackson L, Aljoe H, Dixon RA (2005) Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc Natl Acad Sci 102:16573–16578
Sakagami H, Kushida T, Oizumi T, Nakashima H, Makino T (2010) Distribution of lignin–carbohydrate complex in plant kingdom and its functionality as alternative medicine. Pharmacol Ther 128:91–105
Samuel R, Pu Y, Raman B, Ragauskas AJ (2010) Structural characterization and comparison of switchgrass ball-milled lignin before and after dilute acid pretreatment. Appl Biochem Biotechnol 162:62–74
Shi R, Sun YH, Li Q, Heber S, Sederoff R, Chiang VL (2010) Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol 51:144–163
Stewart JJ, Akiyama T, Chapple C, Ralph J, Mansfield SD (2009) The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar. Plant Physiol 150:621–635
Sudheesh S, Sawbridge TI, Cogan NO, Kennedy P, Forster JW, Kaur S (2015) De novo assembly and characterisation of the field pea transcriptome using RNA-Seq. BMC Genom 16:611
Surget-Groba Y, Montoya-Burgos JI (2010) Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res 20:1432–1440
Sykes RW, Gjersing EL, Foutz K, Rottmann WH, Kuhn SA, Foster CE, Ziebell A, Turner GB, Decker SR, Hinchee MAW, Davis MF (2015) Down-regulation of p-coumaroyl quinate/shikimate 3′-hydroxylase (C3′H) and cinnamate 4-hydroxylase (C4H) genes in the lignin biosynthetic pathway of Eucalyptus urophylla × E. grandis leads to improved sugar release. Biotechnol Biofuels 8:128
Tamasloukht B, Lam MSJWQ, Martinez Y, Tozo K, Barbier O, Jourda C, Jauneau A, Borderies G, Balzergue S, Renou JP, Huguet S, Martinant JP, Tatout C, Lapierre C, Barrière Y, Goffner D, Pichon M (2011) Characterization of a cinnamoyl-CoA reductase 1 (CCR1) mutant in maize: effects on lignification, fibre development, and global gene expression. J Exp Bot 62:3837–3848
Thévenin J, Pollet B, Letarnec B, Saulnier L, Gissot L, Maia-Grondard A, Lapierre C, Jouanin L (2011) The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol Plant 4:70–82
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111
Tu Y, Rochfort S, Liu Z, Ran Y, Griffith M, Badenhorst P, Louie GV, Bowman ME, Smith KF, Noel JP, Mouradov A, Spangenberg G (2010) Functional analyses of caffeic acid O-methyltransferase and cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 22:3357–3373
Upadhyay S, Phukan UJ, Mishra S, Shukla RK (2014) De novo leaf and root transcriptome analysis identified novel genes involved in steroidal sapogenin biosynthesis in Asparagus racemosus. BMC Genom 15:746
Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153:895–905
Vanholme R, Morreel K, Darrah C, Oyarce P, Grabber JH, Ralph J, Boerjan W (2012) Metabolic engineering of novel lignin in biomass crops. New Phytol 196:978–1000
Wang Y, Chantreau M, Sibout R, Hawkins S (2013a) Plant cell wall lignification and monolignol metabolism. Front Plant Sci 4:220
Wang Y, Pan Y, Liu Z, Zhu X, Zhai L, Xu L, Liu L (2013b) De novo transcriptome sequencing of radish (Raphanus sativus L.) and analysis of major genes involved in glucosinolate metabolism. BMC Genom 14:836
Wang GL, Huang Y, Zhang XY, Xu ZS, Wang F, Xiong AS (2016) Transcriptome-based identification of genes revealed differential expression profiles and lignin accumulation during root development in cultivated and wild carrots. Plant Cell Rep 35:1743
Weng JK, Akiyama T, Ralph J, Chapple C (2011) Independent recruitment of an O-methyltransferase for syringyl lignin biosynthesis in Selaginella moellendorffii. Plant Cell 23:2708–2724
Wong MM, Cannon CH, Wickneswari R (2011) Identification of lignin genes and regulatory sequences involved in secondary cell wall formation in Acacia auriculiformis and Acacia mangium via de novo transcriptome sequencing. BMC Genom 12:342
Wu G, Zhang L, Yin Y, Wu J, Yu L, Zhou Y, Li M (2015) Sequencing, de novo assembly and comparative analysis of Raphanus sativus transcriptome. Front Plant Sci 6:198
Yan X, Dong C, Yu J, Liu W, Jiang C, Liu J, Hu Q, Fang X, Wei W (2013) Transcriptome profile analysis of young floral buds of fertile and sterile plants from the self-pollinated offspring of the hybrid between novel restorer line NR1 and Nsa CMS line in Brassica napus. BMC Genom 14:26
Yang L, Wang CC, Guo WD, Li XB, Lu M, Yu CL (2006) Differential expression of cell wall related genes in the elongation zone of rice roots under water deficit. Rus J Plant Physiol 53:390–395
Yates SA, Swain MT, Hegarty MJ, Chernukin I, Lowe M, Allison GG, Ruttink T, Abberton MT, Jenkins G, Skøt L (2014) De novo assembly of red clover transcriptome based on RNA-Seq data provides insight into drought response, gene discovery and marker identification. BMC Genom 15:453
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J (2006) WEGO: a web tool for plotting GO annotations. Nucl Acids Res 34(suppl 2):W293–W297
Yoon J, Choi H, An G (2015) Roles of lignin biosynthesis and regulatory genes in plant development. J Integr Plant Biol 57:902–912
Yu S, Zhang F, Yu Y, Zhang D, Zhao X, Wang W (2012) Transcriptome profiling of dehydration stress in the Chinese Cabbage (Brassica rapa L. ssp. pekinensis) by tag sequencing. Plant Mol Biol Rep 30:17–28
Zhang J, Xiao L, Yang Y, Wang Z, Li G (2014) Lignin binding to pancreatic lipase and its influence on enzymatic activity. Food Chem 149:99–106
Zhang L, Ming R, Zhang J, Tao A, Fang P, Qi J (2015) De novo transcriptome sequence and identification of major bast-related genes involved in cellulose biosynthesis in jute (Corchorus capsularis L.). BMC Genom 16:1062
Acknowledgements
This work was in part supported by grants from the National Key Technology Research and Development Program of China (2017YFD0101803; 2016YFD0100204), Key Technology R & D Program of Jiangsu Province (BE2016379), and Jiangsu Agricultural Science and Technology Innovation Fund (CX(16)1012).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Feng, H., Xu, L., Wang, Y. et al. Identification of critical genes associated with lignin biosynthesis in radish (Raphanus sativus L.) by de novo transcriptome sequencing. Mol Genet Genomics 292, 1151–1163 (2017). https://doi.org/10.1007/s00438-017-1338-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-017-1338-9