Abstract
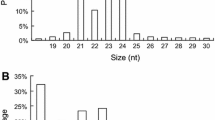
MicroRNAs (miRNAs) are small (20–24 nucleotide) RNAs that are critical regulators of genes involved in diverse plant processes, including development, metabolism, abiotic stress and flowering. Prunus mume is a widely cultivated ornamental plant in East Asia that blooms in early spring, even at temperatures below 0 °C. While miRNAs involved in pistil development have been identified in P. mume, few studies have profiled miRNA expression patterns during flower opening. Here, we used high-throughput sequencing and bioinformatic analysis to identify and profile miRNAs that function during flower opening in P. mume. We identified 47 conserved miRNA sequences belonging to 25 miRNA families from 92 loci in P. mume, along with 33 novel miRNA sequences from 43 loci, including their complementary miRNA* strands. The expression levels of most differentially expressed miRNAs decreased during flower opening, while miR156e-f and miR477b were upregulated at the flowering stage. We predicted 88 target genes for conserved and novel miRNAs using computational analysis and annotated their functions. Seven target genes, encoding squamosa promoter binding protein-like (SPL) and auxin response factor (ARF), scarecrow-like transcription factor (SCL) and APETALA2-like transcription factors (AP2), were verified by 5′-RACE to be the targets of miR156, miR167, miR171 and miR172, respectively. Quantitative real-time PCR validated the expression of the miRNAs and seven target genes. The results help lay the foundation for investigating the roles of miRNAs in the blooming of P. mume.






Similar content being viewed by others
References
Allen E, Xie ZX, Gustafson AM, Carrington JC (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221
An FM, Hsiao SR, Chan MT (2011) Sequencing-based approaches reveal low ambient temperature-responsive and tissue-specific microRNAs in Phalaenopsis orchid. PLoS One 6:e18937
Arenas-Huertero C, Perez B, Rabanal F, Blanco-Melo D, De LRC, Estrada-Navarrete G, Sanchez F, Covarrubias AA, Reyes JL (2009) Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol Biol 70:385–401
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741
Bao N, Lye KW, Barton MK (2004) MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell 7:653–662
Barakat A, Sriram A, Park J, Zhebentyayeva T, Main D, Abbott A (2012) Genome wide identification of chilling responsive microRNAs in Prunus persica. BMC Genomics 13:481
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2011) GenBank. Nucleic Acids Res 39:D32–D37
Bentolila S, Alfonso A, Hanson MR (2002) A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci USA 99:10887–10892
Bernier G, Périlleux C (2005) A physiological overview of the genetics of flowering time control. Plant Biotechnol J 3:3–16
Boss PK, Bastow RM, Mylne JS, Dean C (2004) Multiple pathways in the decision to flower enabling promoting and resetting. Plant Cell 16:18–31
Chen XM (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303:2022–2025
Chen JJ (2012) Study on the flowering process and the postharvest physiological characteristics of Mei flower. Dissertation, Zhejiang A&F University Dissertation
Chen L, Ren YY, Zhang YY, Xu JC, Zhang ZY, Wang YW (2012) Genome-wide profiling of novel and conserved Populus microRNAs involved in pathogen stress response by deep sequencing. Planta 235:873–883
Chuck G, Cigan AM, Saeteurn K, Hake S (2007) The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet 39:544–549
Cordelia B (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218:683–692
Curaba J, Talbot M, Li ZY, Helliwell C (2013) Over-expression of microRNA171 affects phase transitions and floral meristem determinancy in barley. BMC Plant Biol 13:6
Devers E, Branscheid A, May P, Krajinski F (2011) Stars and symbiosis: microRNA-and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol 156:1990–2010
Du DL, Zhang QX, Cheng TR, Pan HT, Yang WR, Sun LD (2012) Genome-wide identification and analysis of late embryogenesis abundant (LEA) genes in Prunus mume. Mol Biol Rep. doi:10.1007/s11033-012-2250-3
Elbashir SM, Lendeekel W, Tusehl T (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 16:1616–1626
Fang Y, You J, Xie K, Xie W, Xiong L (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics 280:535–546
Foucher F, Chevalier M, Corre C, Soufflet-Freslon V, Legeai F, Oyant LH-S (2008) New resources for studying the rose flowering process. Genome 51:827–837
Gao Z, Shi T, Luo X, Zhang Z, Zhuang WB, Wang L (2012) High-throughput sequencing of small RNAs and analysis of differentially expressed microRNAs associated with pistil development in Japanese apricot. BMC Genomics 13:371
Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, Bateman A (2009) Rfam: updates to the RNA families database. Nucleic Acids Res 37:D136–D140
Griffiths-Jones S (2010) miRBase: microRNA sequences and annotation. Curr Protoc Bioinforma Chapter 12: Unit 12.19.11-10
Guddeti S, Zhang DC, Li AL, Leseberg CH, Kang H, Li XG, Zhai WX, Johns MA, Mao L (2005) Molecular evolution of the rice miR395 gene family. Cell Res 15:631–638
Hou JH, Gao ZH, Zhang Z, Chen SM, Ando T, Zhang JY, Wang XW (2011) Isolation and Characterization of an AGAMOUS Homologue PmAG from the Japanese Apricot (Prunus mume Sieb. et Zucc.). Plant Mol Biol Report 29:473–480
Hwang EW, Shin SJ, Yu BK, Byun MO, Kwon HB (2011) miR171 family members are involved in drought response in Solanum tuberosum. J Plant Biol 54:43–48
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Kang M, Zhao Q, Zhu D, Yu J (2012) Characterization of microRNAs expression during maize seed development. BMC Genomics 13:360
Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC (2007) Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5:e57
Kim J, Park JH, Lim CJ, Lim JY, Ryu JY, Lee BW, Choi JP, Kim WB, Lee HY, Choi Y, Kim D, Hur CG, Kim S, Noh YS, Shin C, Kwon SY (2012) Small RNA and transcriptome deep sequencing proffers insight into floral gene regulation in Rosa cultivars. BMC Genomics 13:657
Lertpanyasampatha M, Gao L, Kongsawadworakul P, Viboonjun U, Chrestin H, Liu RY, Chen XM, Narangajavana J (2012) Genome-wide analysis of microRNAs in rubber tree (Hevea brasiliensis L.) using high-throughput sequencing. Planta 236:437–445
Li AL, Long M (2007) Evolution of plant microRNA gene families. Cell Res 17:212–218
Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714
Li B, Qin Y, Duan H, Yin W, Xia X (2011) Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J Exp Bot 62:3765–3779
Liang G, Yang FX, Yu DQ (2010) MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J 62:1046–1057
Lin SI, Santi C, Jobet E, Lacut E, El Kholti N, Karlowski WM, Verdeil JL, Breitler JC, Perin C, Ko SS, Guiderdoni E, Chiou TJ, Echeverria M (2011) Complex regulation of two target genes encoding SPX-MFS proteins by rice miR827 in response to phosphate starvation. Plant Cell Physiol 51:2119–2131
Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AV (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10:50
Lu SF, Sun YH, Shi R, Clark C, Li L, Chiang VL (2005) Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17:2186–2203
Lu SF, Sun YH, Chiang VL (2008) Stress-responsive microRNAs in Populus. Plant J 55:131–151
Luo ZQ (2012) Identification and validation of soybean microRNAs involved in stress responses. Dissertation, Chinese Academy of Agricultural Sciences
Luo ZQ, Jin LG, Qiu LJ (2012) MiR1511 co-regulates with miR1511* to cleave the GmRPL4a gene in soybean. Gene Eng 57:3804–3810
Lv DK, Bai X, Li Y, Ding XD, Ge Y, Cai H, Ji W, Wu N, Zhu YM (2010) Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 459:39–47
Ma HS, Liang D, Shuai P, Xia XL, Yin WL (2010) The salt- and drought inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. J Exp Bot 61:4011–4019
Maher C, Stein L, Ware D (2006) Evolution of Arabidopsis microRNA families through duplication events. Genome Res 16:510–519
Mathews DH, Sabina J, Zuker M, Turner DH (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288:911–940
Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7:e1000148
Meng YJ, Shao CG, Wang HZ, Chen M (2011) The regulatory activities of plant microRNAs: a more dynamic perspective. Plant Physiol 157:1583–1595
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martienssen RA, Poethig RS, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhu JK (2008) Criteria for annotation of plant microRNAs. Plant Cell 20:3186–3190
Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2011) AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819:86–96
Pamp SJ, Frees D, Engelmann S, Hecker M, Ingmer H (2006) Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J Bacteriol 188:4861–4870
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20:3407–3425
Ru P, Xu L, Ma H, Huang H (2006) Plant fertility defects induced by the enhanced expression of microRNA167. Cell Res 16:457–465
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome developmental. Cell 8:517–527
Shi R, Chiang VL (2005) Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39:519–525
Shi T, Gao ZH, Wang LJ, Zhang Z, Zhuang WB, Sun HL, Zhong WJ (2012) Identification of differentially-expressed genes associated with pistil abortion in Japanese apricot by genome-wide transcriptional analysis. PLoS One 7:e47810
Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM (2007) Redundancy and specialization among plant microRNAs: role of the MIR164 family in development robustness. Development 134:1051–1060
Silva C, Garcia-Mas J, Sanchez AM, Arus P, Oliveira MM (2005) Looking into flowering time in almond (Prunus dulcis (Mill) D. A. Webb): the candidate gene approach. Theor Appl Genet 110:959–968
Song QX, Liu YF, Hu XY, Zhang WK, Ma B, Chen SY, Zhang JS (2011) Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol 11:5
Thiebaut F, Rojas CA, Almeida KL, Grativol C, Domiciano GC, Lamb CRC, Engler JDA, Hemerly AS, Ferreira PCG (2012) Regulation of miR319 during cold stress in sugarcane. Plant Cell Environ 35:502–512
Valdes-Lopez O, Yang SS, Aparicio-Fabrc R, Graham PH, Reyes JL, Vance CP, Hernandez G (2010) MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytol 187:805–818
Varkonyi-Gasic E, Gould N, Sandanayaka M, Sutherland P, MacDiarmid RM (2010) Characterisation of microRNAs from apple (Malus domestica ‘Royal Gala’) vascular tissue and phloem sap. BMC Plant Biol 10:159
Wan LC, Zhang HY, Lu SF, Zhang L, Qiu ZB, Zhao YY, Zeng QY, Lin JX (2012) Transcriptome-wide identification and characterization of miRNAs from Pinus densata. BMC Genomics 13:132
Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138:738–749
Wang JW, Park MY, Wang LJ, Koo Y, Chen XY, Weigel D, Poethig RS (2010a) MiRNA control of vegetative phase change in trees. PLoS Genet 7:e1002012
Wang XY, Xiong AS, Yao QH, Zhang Z, Qiao YS (2010b) Direct isolation of high-quality low molecular weight RNA of pear peel. Mol Biotechnol 44:61–65
Wang ZJ, Huang JQ, Huang YJ, Li Z, Zheng BS (2012) Discovery and profiling of novel and conserved microRNAs during flower development in Carya cathayensis via deep sequencing. Planta 236:613–621
Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133:3539–3547
Wu MF, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133:4211–4218
Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138:750–759
Xia R, Zhu H, An YQ, Beers EP, Liu ZR (2012) Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol 13:R47
Xie Y, Sun Y, Li DN, Huang J (2013) MicroRNA828 negatively regulates sucrose-induced anthocyanin biosynthesis in Arabidopsis. Plant Physiol J 49:188–194
Xing S, Salinas M, Hohmann S, Berndtgen R, Huijser P (2010) miR156-targeted and non-targeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22:3935–3950
Yang ZR, Mao X, Yang ZF, Li RZ (2003) Cytochrome P450 genes and their application in plant improvement. HEREDITAS (Beijing) 25:237–240
Yang JH, Han SJ, Yoon EK, Lee WS (2006) Evidence of an auxin signal pathway, microRNA167 -ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res 34:1892–1899
Yang TW, Xue LG, An LZ (2007) Functional diversity of miRNA in plants. Plant Sci 172:423–432
Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC (2011) Widespread regulatory activity of vertebrate microRNA* species. RNA 17:312–326
Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M (2010) Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22:2156–2170
Zanca A, Vicentini R, Ortiz-Morea FA, Bem LED, Silva M, Vincentz M, Nogueira FTS (2010) Identification and expression analysis of microRNAs and targets in the biofuel crop sugarcane. BMC Plant Biol 10:260
Zeng CY, Wang WQ, Zheng Y, Chen X, Bo WP, Song S, Zhang WX, Peng M (2009) Conservation and divergence of microRNAs and their functions in Euphorbiaceous plants. Nucleic Acids Res 38:981–995
Zhang J, Xu Y, Huan Q, Chong K (2009a) Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics 10:449
Zhang Y, Zhang R, Su B (2009b) Diversity and evolution of MicroRNA gene clusters. Sci China Ser C Life Sci 52:261–266
Zhang JH, Zhang SG, Han SY, Wu T, Li XM, Li WF, Qi LW (2012a) Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved microRNAs and their targets during somatic embryogenesis. Planta 236:647–665
Zhang QX, Chen WB, Sun LD, Zhao FY, Huang BQ, Yang WR, Tao Y, Wang J, Yuan ZQ, Fan GY, Xing Z, Han CL, Pan HT, Zhong ZX, Shi WF, Liang XM, Du DL, Sun FM, Xu ZD, Hao RJ, Lv T, Lv YM, Zheng ZQ, Sun M, Luo L, Cai M, Gao YK, Wang JY, Yin Y, Xu X, Cheng TR, Wang J (2012b) The genome of Prunus mume. Nat Commun 3:1318
Zhu QH, Helliwell CA (2011) Regulation of flowering time and floral patterning by miR172. J Exp Bot 62:487–495
Zluvova J, Nicolas M, Berger A, Negrutiu I, Monéger F (2006) Premature arrest of the male flower meristem precedes sexual dimorphism in the dioecious plant Silene latifolia. Proc Natl Acad Sci USA 103:18854–18859
Acknowledgments
This work was supported by the Ministry of Science and Technology (2013AA102607, 2011AA100207).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, T., Pan, H., Wang, J. et al. Identification and profiling of novel and conserved microRNAs during the flower opening process in Prunus mume via deep sequencing. Mol Genet Genomics 289, 169–183 (2014). https://doi.org/10.1007/s00438-013-0800-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-013-0800-6