Abstract
Background
Cancer is a serious public health problem worldwide, and difficulty in early diagnosis has been the chief obstacle to improve the prognosis of patients. Recently, microRNAs (miRNAs) were widely studied to be potential biomarkers for cancer detection. miR-16 is a prevalent but sophisticated one. In the current study, we aimed to assess the diagnostic value of serum miR-16 for cancer detection.
Methods
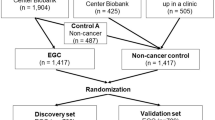
A total of 1458 cancer patients, containing ten types of cancers, and 1457 non-cancer controls were recruited in this study. qRT-PCR was used for the amplification of miRNAs. In addition, a meta-analysis of reported studies was performed to confirm our findings systematically.
Results
Consequently, miR-16 was down-regulated in ESCC, GCA and GNCA patients compared with NCs (all P < 0.001), while up-regulated in PDAC patients (P = 0.001), LAC, LSCC and EEC patients (all P < 0.001). But no significant differences were observed in CRC, EOC and TC patients when compared to NCs (P = 0.747, 0.235 and 0.268, respectively). The areas under the receiver operating characteristic (ROC) curve of miR-16 in GCA, ESCC, LAC, LSCC, GNCA, PDAC and EEC were 0.881, 0.780, 0.757, 0.693, 0.602, 0.614 and 0.681, respectively. Results of meta-analysis showed that miR-16 achieved an overall pooled sensitivity of 0.72, specificity of 0.79, and AUC of 0.85, suggesting that miR-16 was a promising biomarker in cancer detection.
Conclusions
We provided a comprehensive view of the diagnostic value of serum miR-16 in cancer diagnosis, and confirmed that circulating miR-16 could play an important role in cancer detection.




Similar content being viewed by others
References
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6(11):857–866
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99(24):15524–15529
Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM (2005) miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 102(39):13944–13949
El-Abd NE, Fawzy NA, El-Sheikh SM, Soliman ME (2015) Circulating miRNA-122, miRNA-199a, and miRNA-16 as biomarkers for early detection of hepatocellular carcinoma in egyptian patients with chronic hepatitis C virus Infection. Mol Diagn Therapy 19(4):213–220
Ell B, Mercatali L, Ibrahim T, Campbell N, Schwarzenbach H, Pantel K, Amadori D, Kang Y (2013) Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer cell 24(4):542–556
Fan L, Qi H, Teng J, Su B, Chen H, Wang C, Xia Q (2016) Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumour Biol 37(6):7777–7784
Fang C, Zhu DX, Dong HJ, Zhou ZJ, Wang YH, Liu L, Fan L, Miao KR, Liu P, Xu W, Li JY (2012) Serum microRNAs are promising novel biomarkers for diffuse large B cell lymphoma. Ann Hematol 91(4):553–559
Gao L, He SB, Li DC (2014) Effects of miR-16 plus CA19-9 detections on pancreatic cancer diagnostic performance. Clin Lab 60(1):73–77
Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PMM (2003) The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol 56(11):1129–1135
Guo S, Guo W, Li S, Dai W, Zhang N, Zhao T, Wang H, Ma J, Yi X, Ge R, Wang G, Gao T, Li C (2016) Serum miR-16: a potential biomarker for predicting melanoma prognosis. J Investig Dermatol 136(5):985–993
Hanash SM, Baik CS, Kallioniemi O (2011) Emerging molecular biomarkers–blood-based strategies to detect and monitor cancer. Nat Rev Clin Oncol 8(3):142–150
Johansen JS, Calatayud D, Albieri V, Schultz NA, Dehlendorff C, Werner J, Jensen BV, Pfeiffer P, Bojesen SE, Giese N, Nielsen KR, Nielsen SE, Yilmaz M, Hollander NH, Andersen KK (2016) The potential diagnostic value of serum microRNA signature in patients with pancreatic cancer. Int J Cancer 139(10):2312–2324
Ke Y, Zhao W, Xiong J, Cao R (2013) Downregulation of miR-16 promotes growth and motility by targeting HDGF in non-small cell lung cancer cells. FEBS lett 587(18):3153–3157
Li S, Zhang H, Wang X, Qu Y, Duan J, Liu R, Deng T, Ning T, Zhang L, Bai M, Zhou L, Wang X, Ge S, Ying G, Ba Y (2016) Direct targeting of HGF by miR-16 regulates proliferation and migration in gastric cancer. Tumour Biol
Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J, Wang X, Gong Y, Wang W, Kong X (2012) Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer 131(3):683–691
Liu X, Luo HN, Tian WD, Lu J, Li G, Wang L, Zhang B, Liang BJ, Peng XH, Lin SX, Peng Y, Li XP (2013) Diagnostic and prognostic value of plasma microRNA deregulation in nasopharyngeal carcinoma. Cancer Biol Ther 14(12):1133–1142
Maclellan SA, Lawson J, Baik J, Guillaud M, Poh CF, Garnis C (2012) Differential expression of miRNAs in the serum of patients with high-risk oral lesions. Cancer Med 1(2):268–274
Mahn R, Heukamp LC, Rogenhofer S, von Ruecker A, Muller SC, Ellinger J (2011) Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology 77(5):1265 e9–e16
Meng X, Joosse SA, Muller V, Trillsch F, Milde-Langosch K, Mahner S, Geffken M, Pantel K, Schwarzenbach H (2015) Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer 113(9):1358–1366
Min S, Liang X, Zhang M, Zhang Y, Mei S, Liu J, Liu J, Su X, Cao S, Zhong X, Li Y, Sun J, Liu Q, Jiang X, Che Y, Yang R (2013) Multiple tumor-associated microRNAs modulate the survival and longevity of dendritic cells by targeting YWHAZ and Bcl2 signaling pathways. J Immunol 190(5):2437–2446
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105(30):10513–10518
Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M (2011) Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol 45(4):355–360
Ren C, Chen H, Han C, Fu D, Wang D, Shen M (2016) High expression of miR-16 and miR-451 predicating better prognosis in patients with gastric cancer. J Cancer Res Clin Oncol 142(12):2489–2496
Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, Ma X, Han S, Zhang Z (2012) Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci 57(4):897–904
Svoronos AA, Engelman DM, Slack FJ (2016) OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res 76(13):3666–3670
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA 65(2):87–108
Wang H, Wang L, Wu Z, Sun R, Jin H, Ma J, Liu L, Ling R, Yi J, Wang L, Bian J, Chen J, Li N, Yuan S, Yun J (2014) Three dysregulated microRNAs in serum as novel biomarkers for gastric cancer screening. Med Oncol 31(12):298
Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM, Group Q- (2011) QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Int Med 155(8):529–536
Wittmann J, Jack HM (2010) Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta 1806(2):200–207
Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D (2008) miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer 123(2):372–379
Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H, Kou T, Kusaka T, Sasaki Y, Yagi M, Higurashi T, Yukawa N, Amanuma Y, Kikuchi O, Muto M, Ueno Y, Nakajima A, Chiba T, Boland CR, Goel A (2015) Serum miR-21, miR-29a, and miR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin Cancer Res 21(18):4234–4242
Zhang J, Song Y, Zhang C, Zhi X, Fu H, Ma Y, Chen Y, Pan F, Wang K, Ni J, Jin W, He X, Su H, Cui D (2015) Circulating MiR-16-5p and MiR-19b-3p as two novel potential biomarkers to indicate progression of gastric cancer. Theranostics 5(7):733–745
Zhou R, Li X, Hu G, Gong AY, Drescher KM, Chen XM (2012) miR-16 targets transcriptional corepressor SMRT and modulates NF-kappaB-regulated transactivation of interleukin-8 gene. PloS one 7(1):e30772
Zhou X, Zhu W, Li H, Wen W, Cheng W, Wang F, Wu Y, Qi L, Fan Y, Chen Y, Ding Y, Xu J, Qian J, Huang Z, Wang T, Zhu D, Shu Y, Liu P (2015) Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci Rep 5:11251
Zhu C, Ren C, Han J, Ding Y, Du J, Dai N, Dai J, Ma H, Hu Z, Shen H, Xu Y, Jin G (2014) A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer 110(9):2291–2299
Acknowledgements
We thank Ping Liu and Wei Zhu (Department of Oncology, the First Affiliated Hospital of Nanjing Medical University) for constructive suggestion.
Funding
Supported by grants: (1) Wuxi Health and Family Planning Commission Project; Grant number: Q201724. (2) Kunshan Social Development Science and Technology Project; Grant number: KS1643.
Author information
Authors and Affiliations
Contributions
HZB, CWJ and DYP performed experiments; HZB conducted the meta-analysis; GQ and SD reviewed eligible articles; WXH and MY provided the serum samples of patients and normal controls; HZB and ZX drafted the manuscript; ZX and HD designed the study, critically interpreted results; All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethics approval
All the procedures were approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University and the Affiliated Hospital of Jiangnan University in compliance with the Declaration of Helsinki, and the written informed consent was obtained from each participant.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1 Risk of bias and applicability concerns graph a review of authors’ judgments about each domain presented as percentages across included studies.
Figure S2 Forest plots of pooled A: positive likelihood ratio (PLR); B: negative likelihood ratio (NLR); C: diagnostic odds ratio (DOR) for miR-16 in cancer diagnosis.
Rights and permissions
About this article
Cite this article
Huang, Z., Chen, W., Du, Y. et al. Serum miR-16 as a potential biomarker for human cancer diagnosis: results from a large-scale population. J Cancer Res Clin Oncol 145, 787–796 (2019). https://doi.org/10.1007/s00432-019-02849-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-019-02849-8