Abstract
Main conclusion
An efficient method of DNA-free gene-editing in potato protoplasts was developed using linearized DNA fragments, UBIQUITIN10 promoters of several plant species, kanamycin selection, and transient overexpression of the BABYBOOM transcription factor.
Abstract
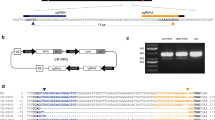
Plant protoplasts represent a reliable experimental system for the genetic manipulation of desired traits using gene editing. Nevertheless, the selection and regeneration of mutated protoplasts are challenging and subsequent recovery of successfully edited plants is a significant bottleneck in advanced plant breeding technologies. In an effort to alleviate the obstacles related to protoplasts’ transgene expression and protoplasts’ regeneration, a new method was developed. In so doing, it was shown that linearized DNA could efficiently transfect potato protoplasts and that UBIQUITIN10 promoters from various plants could direct transgene expression in an effective manner. Also, the inhibitory concentration of kanamycin was standardized for transfected protoplasts, and the NEOMYCIN PHOSPHOTRANSFERASE2 (NPT2) gene could be used as a potent selection marker for the enrichment of transfected protoplasts. Furthermore, transient expression of the BABYBOOM (BBM) transcription factor promoted the regeneration of protoplast-derived calli. Together, these methods significantly increased the selection for protoplasts that displayed high transgene expression, and thereby significantly increased the rate of gene editing events in protoplast-derived calli to 95%. The method developed in this study facilitated gene-editing in tetraploid potato plants and opened the way to sophisticated genetic manipulation in polyploid organisms.








Similar content being viewed by others
Data availability
The data underlying this article are available in the article and in its online supplemental material.
Abbreviations
- BBM:
-
BABYBOOM
- GB:
-
GoldenBraid
- GMO:
-
Genetically modified organisms
- NPT2:
-
NEOMYCIN PHOSPHOTRANSFERASE 2
- TALEN:
-
Transcription activator-like effector nucleases
- YFP:
-
Yellow fluorescent protein
References
Andersson M, Turesson H, Nicolia A et al (2017) Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep 36:117–128
Andersson M, Turesson H, Olsson N et al (2018) Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol Plant 164:378–384
Ballas N, Zakai N, Friedberg D, Loyter A (1988) Linear forms of plasmid DNA are superior to supercoiled structures as active templates for gene expression in plant protoplasts. Plant Mol Biol 11:517–527
Barrell PJ, Meiyalaghan S, Jacobs JME, Conner AJ (2013) Applications of biotechnology and genomics in potato improvement. Plant Biotechnol J 11:907–920
Benfey PN, Chua NH (1990) The Cauliflower Mosaic Virus 35S Promoter: combinatorial regulation of transcription in plants. Science 250:959–966
Bouchabké-Coussa O, Obellianne M, Linderme D et al (2013) Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep 32:675–686
Boutilier K, Offringa R, Sharma VK et al (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14:1737–1749
Burris KP, Dlugosz EM, Collins AG et al (2016) Development of a rapid, low-cost protoplast transfection system for switchgrass (Panicum virgatum L.). Plant Cell Rep 35:693–704
Che P, Wu E, Simon MK et al (2021) Wuschel2 enables highly efficient CRISPR/Cas-targeted genome editing during rapid de novo shoot regeneration in sorghum. bioRxiv. https://doi.org/10.1038/s42003-022-03308-w
Chen C, Fu X, Peng R et al (2020) Detoxifying processes during kanamycin-induced stress to Arabidopsis thaliana seedling growth. Biotechnol Biotechnol Equip 34:673–679
Chupeau M-C, Granier F, Pichon O et al (2013) Characterization of the early events leading to totipotency in an Arabidopsis protoplast liquid culture by temporal transcript profiling. Plant Cell 25:2444–2463
Clasen BM, Stoddard TJ, Luo S et al (2016) Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol J 14:169–176
Dalla Costa L, Piazza S, Pompili V et al (2020) Strategies to produce T-DNA free CRISPRed fruit trees via Agrobacterium tumefaciens stable gene transfer. Sci Rep 10:20155
Davey MR, Anthony P, Power JB, Lowe KC (2005) Plant protoplasts: status and biotechnological perspectives. Biotechnol Adv 23:131–171
Domínguez A, Cervera M, Pérez RM et al (2004) Characterisation of regenerants obtained under selective conditions after Agrobacterium-mediated transformation of citrus explants reveals production of silenced and chimeric plants at unexpected high frequencies. Mol Breed 14:171–183
Doyle J (1991) DNA protocols for plants. In: Hewitt GM, Johnston AWB, Young JPW (eds) Molecular techniques in taxonomy. Springer, Berlin Heidelberg, pp 283–293
González MN, Massa GA, Andersson M et al (2019) Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front Plant Sci 10:1649
Gordon-Kamm B, Sardesai N, Arling M et al (2019) Using morphogenic genes to improve recovery and regeneration of transgenic plants. Plants 8:38. https://doi.org/10.3390/plants8020038
Grefen C, Donald N, Hashimoto K et al (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64:355–365
Hensel G, Oleszczuk S, Daghma DES et al (2012) Analysis of T-DNA integration and generative segregation in transgenic winter triticale (x Triticosecale Wittmack). BMC Plant Biol 12:171
Horstman A, Bemer M, Boutilier K (2017a) A transcriptional view on somatic embryogenesis. Regeneration (oxf) 4:201–216
Horstman A, Li M, Heidmann I et al (2017b) The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol 175:848–857. https://doi.org/10.1104/pp.17.00232
Hsu C-T, Yuan Y-H, Zheng P-X et al (2021) DNA-free CRISPR-Cas9 gene editing of tetraploid tomatoes using protoplast regeneration. bioRxiv. https://doi.org/10.1093/plphys/kiac022
Hu Y, Song D, Gao L et al (2020) Optimization of isolation and transfection conditions of maize endosperm protoplasts. Plant Methods 16:96
Jeong YY, Lee H-Y, Kim SW et al (2021) Optimization of protoplast regeneration in the model plant Arabidopsis thaliana. Plant Methods 17:21
Jiang P, Zhang K, Ding Z et al (2018) Characterization of a strong and constitutive promoter from the Arabidopsis serine carboxypeptidase-like gene AtSCPL30 as a potential tool for crop transgenic breeding. BMC Biotechnol 18:59
Kumar M, Ayzenshtat D, Marko A, Bocobza S (2021) Optimization of T-DNA configuration with UBIQUITIN10 promoters and tRNA-sgRNA complexes promotes highly efficient genome editing in allotetraploid tobacco. Plant Cell Rep 41:175–194. https://doi.org/10.1007/s00299-021-02796-0
Li J-F, Norville JE, Aach J et al (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691
Li X, Sandgrind S, Moss O et al (2021) Efficient protoplast regeneration protocol and CRISPR/Cas9-mediated editing of glucosinolate transporter (GTR) genes in rapeseed (Brassica napus L.). Front Plant Sci 12:680859
Lin C-S, Hsu C-T, Yang L-H et al (2018) Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol J 16:1295–1310
Lowe K, Wu E, Wang N et al (2016) Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28:1198–2015. https://doi.org/10.1105/tpc.16.00124
Lowe K, La Rota M, Hoerster G et al (2018) Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell Dev Biol Plant 54:240–252
Mao Y, Botella JR, Liu Y, Zhu J-K (2019) Gene editing in plants: progress and challenges. Natl Sci Rev 6:421–437
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Nadakuduti SS, Buell CR, Voytas DF et al (2018) Genome editing for crop improvement—applications in clonally propagated polyploids with a focus on potato (Solanum tuberosum L.). Front Plant Sci 9:1607
Nicolia A, Proux-Wéra E, Åhman I et al (2015) Targeted gene mutation in tetraploid potato through transient TALEN expression in protoplasts. J Biotechnol 204:17–24
Nicolia A, Andersson M, Hofvander P et al (2021) Tomato protoplasts as cell target for ribonucleoprotein (RNP)-mediated multiplexed genome editing. Plant Cell Tissue Organ Cult 144:463–467
Passarinho P, Ketelaar T, Xing M et al (2008) BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol Biol 68:225–237
Potenza C, Aleman L, Sengupta-Gopalan C (2004) Targeting transgene expression in research, agricultural, and environmental applications: promoters used in plant transformation. In Vitro Cell Dev Biol Plant 40:1–22
Reed KM, Bargmann BOR (2021) Protoplast regeneration and its use in new plant breeding technologies. Front Genome Ed 3:734951
Salam BB, Barbier F, Danieli R et al (2021) Sucrose promotes stem branching through cytokinin. Plant Physiol 185:1708–1721
Sandgrind S, Li X, Ivarson E et al (2021) Establishment of an efficient protoplast regeneration and transfection protocol for field cress (Lepidium campestre). Front Genome Ed 3:757540
Sarrion-Perdigones A, Vazquez-Vilar M, Palací J et al (2013) GoldenBraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol 162:1618–1631
Srinivasan C, Liu Z, Heidmann I et al (2007) Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.). Planta 225:341–351
Waibel F, Filipowicz W (1990) U6 snRNA genes of Arabidopsis are transcribed by RNA polymerase III but contain the same two upstream promoter elements as RNA polymerase II-transcribed U-snRNA genes. Nucleic Acids Res 18:3451–3458
Wang X, Xu L, Liu X et al (2021) Development of potent promoters that drive the efficient expression of genes in apple protoplasts. Hortic Res 8:211
Wang J, Wang Y, Lü T et al (2022) An efficient and universal protoplast isolation protocol suitable for transient gene expression analysis and single-cell RNA sequencing. Int J Mol Sci 23:3419. https://doi.org/10.3390/ijms23073419
Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112:3570–3575
Xu M, Du Q, Tian C et al (2021) Stochastic gene expression drives mesophyll protoplast regeneration. Sci Adv. https://doi.org/10.1126/sciadv.abg8466
Zhang T-Q, Lian H, Zhou C-M et al (2017) A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29:1073–1087
Zhao X, Jayarathna S, Turesson H et al (2021) Amylose starch with no detectable branching developed through DNA-free CRISPR-Cas9 mediated mutagenesis of two starch branching enzymes in potato. Sci Rep 11:4311
Acknowledgements
We are very thankful to Dr. Tzahi Arazi for his advice and critical reading of this manuscript. We are grateful to Per Hofvander and Mariette Andersson for their assistance in achieving regeneration from protoplast. We also thank Amit Gal-on and Diana Leibman for providing the pSAT vector used to knockout StVINV.
Funding
This study was financially supported by the Chief Scientist—Ministry of Agriculture and Rural Development NO. 20-01-0209 as part of the National Center for Genome Editing in Agriculture.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rather, G.A., Ayzenshtat, D., Teper-Bamnolker, P. et al. Advances in protoplast transfection promote efficient CRISPR/Cas9-mediated genome editing in tetraploid potato. Planta 256, 14 (2022). https://doi.org/10.1007/s00425-022-03933-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-03933-z