Abstract
Although the hormones, gibberellin and auxin, are known to play a role in the initiation of fruits, no such function has yet been demonstrated for abscisic acid (ABA). However, ABA signaling and ABA responses are high in tomato (Solanum lycopersicum L.) ovaries before pollination and decrease thereafter (Vriezen et al. in New Phytol 177:60–76, 2008). As a first step to understanding the role of ABA in ovary development and fruit set in tomato, we analyzed ABA content and the expression of genes involved in its metabolism in relation to pollination. We show that ABA levels are relatively high in mature ovaries and decrease directly after pollination, while an increase in the ABA metabolite dihydrophaseic acid was measured. An important regulator of ABA biosynthesis in tomato is 9-cis-epoxy-carotenoid dioxygenase (LeNCED1), whose mRNA level in ovaries is reduced after pollination. The increased catabolism is likely caused by strong induction of one of four newly identified putative (+)ABA 8′-hydroxylase genes. This gene was named SlCYP707A1 and is expressed specifically in ovules and placenta. Transgenic plants, overexpressing SlCYP707A1, have reduced ABA levels and exhibit ABA-deficient phenotypes suggesting that this gene encodes a functional ABA 8′-hydroxylase. Gibberellin and auxin application have different effects on the LeNCED1 and SlCYP707A1 gene expression. The crosstalk between auxins, gibberellins and ABA during fruit set is discussed.
Similar content being viewed by others
Introduction
A major and nutritionally important part of the human diet is composed of fruits or fruit-derived products. In addition, fruits are important in the life cycle of plants, since they protect the seeds during their development and allow seed dispersal. Therefore, there has been considerable research on fruit development, for which tomato has been used as a model system for physiological and molecular studies for a long time. The first step of fruit development, fruit initiation, is very important from an agro-economical point of view. It is a very delicate phase that is sensitive to abiotic factors, which often decrease fruit set and thus reduce yields. From a developmental point of view, fruit initiation, commonly referred to as fruit set, is the stage at which the ovary makes the decision to abort or to proceed with further cell division and fruit development (Gillaspy et al. 1993). Normally, fruit set is dependent on successful completion of pollination and fertilization (Gillaspy et al. 1993). However, it is not known precisely how the signals from pollination and fertilization stimulate cell division and fruit initiation.
It has long been known that application of hormones such as auxins or gibberellins can induce fruit set in the absence of fertilization. The resulting parthenocarpic fruit growth is a clear demonstration of the importance of hormones in fruit initiation and development. There are a number of parthenocarpic tomato mutants, such as pat, pat2 and pat3/4, in which it was shown that the GA content and/or GA biosynthesis were increased significantly (Mazzucato et al. 1998; Fos et al. 2000, 2001; Olimpieri et al. 2007). Furthermore, overexpression of the iaaM gene, which encodes an enzyme involved in auxin biosynthesis, also induced parthenocarpic fruit development in tomato and many other species (Rotino et al. 1997; Mezzetti et al. 2004). Changes in both GA and auxin content were thus associated with parthenocarpic phenotypes. The altered hormone balance in the ovary of parthenocarpic plants seems, therefore, to substitute for pollination and fertilization (Gorguet et al. 2005).
Some of the genes functioning in the biosynthesis and signaling cascade of these hormones during fruit initiation have been identified recently. For instance, it was shown that expression of a GA 20-oxidase gene was induced by pollination in tomato (Rebers et al. 1999; Serrani et al. 2007), and the importance of SlDELLA, which represses GA signaling and tomato fruit set, has also been demonstrated (Marti et al. 2007). In additional, IAA9 and ARF8, both transcriptional regulators of the auxin response, were shown to be part of an important signal for fruit set (Wang et al. 2005; Goetz et al. 2007). However, many other genes remain to be discovered and investigated. We have used transcript profiling to gain a better insight into the genes involved in fruit initiation, focusing on genes involved in hormonal signaling. Several groups of genes were found to have altered mRNA levels after pollination including genes involved in the cell cycle, posttranscriptional gene regulation, GA biosynthesis and auxin signaling (Vriezen et al. 2008). Notably, several genes related to abscisic acid (ABA), such as ABA RESPONSE ELEMENT BINDING PROTEIN 1, ABA INSENSITIVE1-like genes, and dehydrin genes, were found to be highly expressed in mature ovaries while their expression decreased after pollination (Vriezen et al. 2008). Many factors related to ABA signaling and ABA responses are thus higher in mature, unpollinated ovaries and are quickly reduced after pollination. ABA is known to play a role in seed and bud dormancy, and in the regulation of abiotic stress responses (Horvath et al. 2003; Bartels and Sunkar 2005; Finkelstein et al. 2008). During tomato fruit development, its role is thought to be primarily restricted to the latter half of seed development in processes such as reserve deposition, desiccation tolerance and seed dormancy (Gillaspy et al. 1993), and a role in the first phase of fruit initiation has not been established. ABA has, however, been mentioned as a player in pea fruit set (Garcia-Martinez and Carbonell 1980; Rodrigo and Garcia-Martinez 1998). Here, we describe the changes found in the levels of ABA and the ABA metabolites dihydrophaseic acid (DPA) and phaseic acid (PA) in tomato ovaries and the regulation of ABA biosynthesis and catabolism genes during fruit initiation. Our data suggest that ABA is an additional player in the regulation of tomato fruit set together with auxin and GA.
Materials and methods
Plant material
Tomato plants (Solanum lycopersicum L. cv. Moneymaker from Enza Zaden, Enkhuizen, The Netherlands) were grown under greenhouse conditions from March to October under a 16/8 h day–night rhythm. Supplementary lights (600 W high-pressure sodium lights) turned on below 200 W/m2 and turned off above 300 W/m2. Temperature was kept above 20°C during the light period and 17°C during the dark period with the PRIVA Integro versie 724 system. Plants were watered daily and given fertilizer weekly.
Treatments
Flowers were emasculated 3 days before full bloom (dbf) and at full bloom either hand pollinated or treated with 2 μL of either 1 mM GA3 (Duchefa Biochemie BV, Haarlem, The Netherlands) in 1% (v/v) ethanol or 1 mM 4-Cl-IAA (Sigma-Aldrich, St. Louis, MO, USA) in 2% (v/v) ethanol, or 1 mM ABA (Acros, Geel, Belgium) in 10% (v/v) methanol. 4-Cl-IAA is a more stable form of auxin than IAA and was used because it appeared to induce tomato fruit set more reliably than IAA. Control flowers were treated with 2 μL of the solvent (10% methanol or 1% ethanol). Hormone treatments were repeated after 2 days. Flowers were harvested 3 days after first treatment unless otherwise noted. Plant material was sampled between 1100 and 1300 hours and directly frozen in liquid nitrogen. When necessary, plant material was dissected using binoculars and frozen thereafter in liquid nitrogen. The different tissues were dissected from young flowers that were about 3 dbf. Roots and hypocotyls were isolated from 10-day-old seedlings.
ABA analysis
Abscisic acid was quantified in triplicates by GC–MS as described for GA analysis (Griffiths et al. 2006), but with modifications. Freeze-dried samples were homogenized in 50 mL 80% (v/v) methanol/water in 100 mL flasks after which 300 ng 3-methyl-[2H3]ABA was added as internal standard. Samples were purified as described previously except that the pooled ethyl acetate phases after elution from the Varian Bond Elut NH2 cartridge (100 mg; Kinesis, St. Neots, UK) were evaporated to dryness in vacuo, and dissolved in ethyl acetate (20 μL) of which 2 μL was injected into a TR-1 capillary column (30 m × 0.25 mm × 0.25 mm film thickness; Thermo Fisher Scientific, Madison, WI, USA) at 50°C. The split valve (50:1) was opened after 2 min and the temperature increased at 20°C/min to 180°C and then at 4°C/min to 300°C. The instrument was operated in selective ion monitoring mode, monitoring the ions at m/z 190 and 162 for ABA and 193 and 166 for [2H3]ABA. The amount of ABA was determined from the peak areas for the ions m/z 190 and 193 by reference to a calibration curve.
ABA catabolite analysis
The ABA catabolite analysis was performed at the Plant Biotechnology Institute of the National Research Council of Canada (http://www.pbi.nrc.gc.ca/ENGLISH/technology-platforms/plant-hormone-profiling.htm) by high-performance liquid chromatography electrospray tandem mass spectrometry (HPLC-ES-MS/MS) using deuterated internal standards, as described in Owen and Abrams (2009).
Neighbor joining tree
NCED mRNA sequences from Arabidopsis, rice, maize, tomato and potato were obtained from NCBI (www.ncbi.nlm.nih.gov) or Dana-Farber Cancer Institute (DFCI) Tomato Gene Index (http://compbio.dfci.harvard.edu/tgi). The sequences were translated to protein and aligned with ClustalW (www.ebi.ac.uk/clustalw/). A neighbor joining tree was generated using the PHYLIP Protdist program (Phylogeny Inference Package version 3.5c; J. Felsenstein, Department of Genetics, University of Washington, Seattle) available on http://bioweb.pasteur.fr/seqanal/phylogeny/phylip-uk.html. Default parameters were used and 100 bootstrap replicates were generated. The consensus tree was drawn in TreeView (free available from http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). An unrooted tree was drawn from output data without bootstrap values, bootstrap values (calculated by Prodist) were manually added afterwards.
Isolation and cloning of SlCYP707A1 and CYP707 like cDNAs
The full-length SlCYP707A1 sequence was isolated by plaque screening of a phage cDNA library (HybriZAP® 2.1, Stratagene, La Jolla, CA, USA) with a probe corresponding to base 1,024–1,387 of SlCYP707A1, following the manufacturer’s protocol. TC177455, AI484420 and TC186477 sequences were obtained from the DFCI Tomato Gene Index.
RNA isolation, cDNA synthesis and Q-PCR data analysis
RNA was isolated with the RNAeasy kit (Qiagen, Valencia, CA, USA). Photometric RNA measurements were done to equilibrate the RNA concentrations of different samples. Equal amounts of RNA were DNase treated (RQ1, Promega, Madison, WI, USA). RNA (0.5 μg) was reverse transcribed (RT) in a total volume of 10 μL using a cDNA synthesis kit (iScript™, Bio-Rad Laboratories, Hercules, CA, USA) following manufacturer’s protocol.
Real-time quantitative PCR (Q-PCR) primers were designed using a computer program (Beacon Designer Software, Premier Biosoft International, CA, USA). Alignments of genes that were part of a gene family were made and gene specific DNA pieces were used for primer design. Primer pairs are depicted in Table 1. Specificity was tested by sequencing the amplicon. Q-PCRs were done using SYBR green mix (iQ-SYBR Green Supermix, Bio-Rad Laboratories). PCR reactions were performed in a 96-well thermocycler (Bio-Rad iCycler) using a temperature program starting with 3 min at 95°C then 40 cycles consisting of 15 s at 95°C and 45 s at 57°C and finally the melting temperature of the amplified product was determined to verify the presence of a single product. Five microliters of 25-fold diluted cDNA was used per sample. Technical and biological replicates were always performed. Both actin 2/7 and ubiquitin 7 were used as internal control genes, to correct for difference in cDNA amounts. Diluted DNase-treated RNA was also included in the Q-PCR as a control for genomic DNA contamination.
Relative mRNA levels were calculated following the Bio-Rad outlined methodology based on Vandesomple et al. (2002) and corrected for PCR efficiencies, which were determined using dilution ranges (0.1–10 pg/μL). The average of two biological repeats and two technical repeats is depicted together with the SE. When error bars were omitted in graphs, biological replicates showed the same trend but the absolute amounts were different—in these cases, only one biological replicate is shown.
Semi-quantitative PCR
Semi-quantitative PCRs were performed using 5 μL of 25-fold diluted cDNA, buffer IV, 2.5 mM MgCl2, 0.5 unit Red Hot Taq DNA polymerase (all from ABgene Limited, Epson, Surrey, UK), 0.4 mM dNTPs (Fermentas, St. Leon-Rot, Germany), and 0.1 μM primers (Table 1) in a reaction of 30 cycles, each comprising of 15 s at 95°C (denaturation), 30 s at 57°C (primer annealing) and 10 s at 72°C (extension time).
In situ hybridization
The RNA probes were synthesized by transcribing bases 1,467–1,648 of the CYP707A1 cDNA using T7 (sense) and SP6 (anti-sense) RNA polymerase and digoxigenin (DIG)-labeled UTP (Roche Applied Science, Basel, Switzerland). DIG labeling efficiencies were tested by a spot assay. Tissue fixation was done as described by Bereterbide et al. (2002). Embedded tissues were sliced into 8 μm sections. Paraplast Plus (Sigma-Aldrich, St. Louis, MO, USA) was removed with Histoclear (National Diagnostics, Atlanta, GA, USA), sections were hydrated, immersed in TE (Tris–HCl pH 7.5, 10 mM EDTA) and a proteinase K treatment was performed. Sections were then dehydrated again, and air dried for 2–3 h. The slides were incubated in a humidified box overnight at 50°C, in 150 μL hybridization mixture (50% formamide, 0.5 μg/mL tRNA, 10% (w/v) dextran sulfate, 300 mM NaCl, 5 mM EDTA, 10 mM Tris–HCl pH 7.5) supplemented with 100 ng of probe. The next morning slides were immersed in a series of SSC (Na3citrate, NaCl) washing buffers until 0.5× SSC. Anti-DIG alkaline phosphatase-coupled antibody (Roche Applied Science) was diluted 500-fold in 1% (w/v) BSA solution and after blocking a volume of 150 μL was added to the slides and incubated in a humid chamber at 37°C for 1.5 h. After two washes, the slides were put in color substrate reaction buffer [100 mM NaCl, 5 mM MgCl2, 100 mM Tris–HCl pH 9.0, 10% (w/v) polyvinylalcohol, 0.24 mg/mL Levamisole (Sigma-Aldrich), 5 μL/mL NBT and 3.75 μL/mL BCIP (Roche Applied Science)] in the dark until purple staining appeared. Staining was stopped by immersing the slides in TE.
Overexpression lines
To generate transgenic overexpression SlCYP707A1 lines, the coding region (base 6–1,478) was PCR amplified and cloned in the Gateway entry vector pENTR/D-TOPO vector (Invitrogen, Carlsbad, CA, USA). SlCYP707A1 coding region was recombined between the Cauliflower Mosaic Virus 35S promoter in the pGD625 vector (Chalfun-Junior et al. 2005) and the NOPALINE SYNTHASE terminator. Transgenic plants were generated by Agrobacterium tumefaciens-mediated transformation. Cotyledons of 10-day-old seedlings were cut and pre-incubated overnight on co-cultivation MS medium containing vitamin Gamborg B5, 1% (v/w) sucrose, 0.5 mg/L Mes buffer, 0.8% (v/w) Daishin agar, 0.05 mg/L 2,4-D, 0.1 mg/L IAA, 2.0 mg/L zeatin, 200 μM acetosyringone, pH 5.8. A. tumefaciens (EHA 105) were grown to an OD600 of 1.0 and diluted 25 times in liquid LB media and grown for another 4 h to an OD600 between 0.2 and 0.3. Bacteria were pelleted and resuspended in liquid MS medium containing vitamin Gamborg B5 mixture, 3% (w/v) sucrose, 0.5 mg/L Mes buffer, 200 μM acetosyringone. Cotyledons were incubated 30 min in the bacterial suspension, washed and placed back on the co-cultivation media. Two days after transformation, cotyledons were placed on MS medium containing vitamin Gamborg B5 mixture, 3% (v/w) sucrose, 0.5 mg/L Mes buffer, 0.8% (v/w) Daishin agar, 0.1 mg/L IAA, 2.0 mg/L zeatin, 200 mg/L cefotaxime and 50 mg/L vancomycin and incubated for another 2 days. To induce callus and shoot formation, cotyledons were transferred to shoot inducing MS medium containing vitamin Gamborg B5 mixture, 1% (w/v) glucose, 0.5 mg/L Mes buffer, 0.8% (w/v) Daishin agar, 0.1 mg/L IAA, 2.0 mg/L zeatin, 500 mg/L carbenicillin and 100 mg/L kanamycin. Cotyledons were transferred to fresh medium every 3 weeks. Elongated shoots of 2–4 cm were excised from the callus and transferred to rooting MS culture medium containing 1.5% (w/v) sucrose, 4% (w/v) purified agar (Oxoid Ltd, Hampshire, UK), 0.25 mg/L IBA, 100 mg/L cefotaxime and 100 mg/L vancomycin. Rooted plantlets were transferred to soil for further selection including, ploidy test and PCR with primers specific for the kanamycin resistance gene. Media components and antibiotics were obtained from Duchefa Biochemie BV unless otherwise specified.
Water stress experiment
Pots of wild type and SlCYP707A1 overexpression lines of the same age and size were saturated with water at the start of the experiment. Plants were withheld water from then on. Photographs were taken 72 h after start of the experiment.
Leaf surface area
Leaf surface area was measured for four compound leaves of 2 months old tomato plants on a Li-3100 area meter (Li-Cor, Lincoln, NE, USA). Average leaf surface is depicted with SD. Leafs were always picked at the same height.
Accession details
GenBank accession numbers: LeNCED1 (Z97215), AtNCED2 (NM117945), AtNCED3 (NM112304), AtNCED4 (NM118036), AtNCED5 (NM102749), AtNCED6 (NM113327), AtNCED9 (NM106486), StNCED1 (AY662343), OsNCED1 (AY838897), OSNCED2 (AY838898), OsNCED3 (AY838899), OsNCED4 (AY838900), OsNCED5 (AY838901), VP14 (ZMU95953), AhNCED1 (AJ574819), SlCYP707A1 (EU183406), SlCYP707A3 (AI484420), StCYP707A2 (DQ206631), AtCYP707A1 (NM202845), AtCYP707A2 (NM128466), AtCYP707A3 (NM180805), AtCYP707A4 (NM112814), ACT2/7 (BT013707), UBQ7 (AK246454).
DFCI Tomato Gene Index accession numbers: TC175556, TC154638, TC154637, TC156234, SlCYP707A2 (TC177455), SlCYP707A4 (TC186477).
Results
ABA content
Abscisic acid signaling and ABA response genes are highly expressed in unpollinated ovaries and their expression decreases upon pollination (Vriezen et al. 2008), suggesting a concomitant decrease in ABA concentration. To confirm this assumption, ABA concentration was determined in ovaries at several time points before and after pollination. Figure 1 shows that the free-ABA level in ovaries significantly decreased from 6.7 ng/mg DW before pollination (C0) to 2.4 ng/mg DW 3 days after pollination (dap). In contrast, if ovaries were kept unpollinated during the same period, the free-ABA level increased to 7.6 ng/mg DW (C3).
Regulation of ABA biosynthesis
It is generally accepted that the cleavage reaction catalyzed by 9-cis-epoxy-carotenoid dioxygenases (NCED) is a major rate-limiting step and point of regulation in ABA biosynthesis (Qin and Zeevaart 2002). In other species, such as Arabidopsis and rice, NCED enzymes are encoded by small gene families composed of five and three members, respectively (Tan et al. 2003). However, in tomato only one NCED (LeNCED1) has been identified so far, and shown to have 9-cis-epoxy-carotenoid cleavage activity. In an attempt to obtain a complete overview of the transcriptional regulation of the whole NCED gene family in tomato, we searched the DFCI Tomato Gene Index and NCBI EST databases for sequences homologous to LeNCED1 or to one of the five known Arabidopsis genes. The neighbor joining tree in Fig. 2 shows that the four tomato sequences most similar to LeNCED1 did not group together with the LeNCED1 and the other functional NCED genes from Arabidopsis, rice, potato and maize. Three of the four tomato sequences (TC175556, TC154638, and TC154637) formed a separate group, while the other sequence (TC156234) grouped together with other carotenoid cleavage dioxygenases (CCDs) that are not known to be related to ABA biosynthesis (Tan et al. 2003). In addition, the deduced amino acid sequences of the four tomato sequences were less than 35% identical to the functional Arabidopsis or tomato NCED amino acid sequences. Therefore, we think it is highly likely that no additional functional NCED genes are present in the tomato genome.
Neighbor joining tree of NCED and NCED-like sequences with bootstrap values indicated at junctions. LeNCED1 groups together with known NCED sequences such as AtNCED2,3,5,6,9, VP14 and OsNCED3,5,6. Two other groups are formed including TC175556, TC154638, TC154637, TC156234, OsNCED1,2 and AtNCED4. Tomato sequences are underlined. For accession details see “Materials and methods”
LeNCED1 mRNA levels decreased after pollination in both the ovules and placenta tissue and the pericarp (Fig. 3a). Semi-quantitative PCR analysis, with specific LeNCED1 primers, revealed that it is expressed equally in wall, ovules and placenta in unpollinated ovaries (Fig. 4a). After pollination, there is a decrease in expression in wall and ovules. Because of its equal distribution throughout the unpollinated ovary, no in situ hybridization was performed for LeNCED1.
a Relative mRNA levels of LeNCED1. LeNCED1 mRNA levels decrease in both pericarp and ovules/placenta after pollination. b mRNA levels of SlCYP707A1, SlCYP707A2, SlCYP707A3, and SlCYP707A4 (CYP1–CYP4) in ovaries 3 days after full bloom (control) or after pollination. c mRNA levels of SlCYP707A1 in different generative and vegetative tissues of tomato; hypocotyl (H) and root (R) tissue from 10-day-old seedlings, and pedicel (P), sepal (Se), petal (Pt), anther (A) and ovary (O) tissue from flowers 3 days before full bloom. d mRNA levels of SlCYP707A1 in more detail in separated ovules/placenta and pericarp tissue. SlCYP707A1 mRNA levels are increased after pollination specifically in ovule/placenta tissue. Mean values (n = 4) ± SE
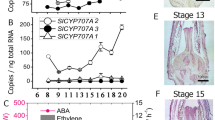
Localization of LeNCED1 and SlCYP707A1 mRNA in ovaries. a Semi-quantitative PCR with LeNCED1 and SlCYP707A1 specific primers, and actin 2/7 as an internal control gene. LeNCED1 is equally expressed in wall, placenta and ovules of unpollinated ovaries and expressed lower after pollination in wall and ovules. SlCYP707A1 is weakly expressed in the placenta of unpollinated ovaries and expressed higher in both ovules and placenta in pollinated ovaries. In situ hybridization with a sense (b) and anti-sense (c) RNA probe of SlCYP707A1. Alkaline phosphatase activity gives a purple staining. Bar represents 100 μm
Regulation of ABA catabolism
Abscisic acid concentration is regulated by catabolism as well as synthesis. In plant cells, ABA is mainly inactivated by 8′-hydroxylation (Nambara and Marion-Poll 2005), which is in Arabidopsis catalyzed by four cytochrome P450 mono-oxygenases, AtCYP707A1 to AtCYP707A4 (Kushiro et al. 2004). We previously identified a tomato homolog of AtCYP707A4, which was strongly induced in the ovary after pollination (Vriezen et al. 2008). We have now isolated and characterized the corresponding full-length (1,742 bp) cDNA clone from a tomato ovary cDNA library. The deduced amino acid sequence is highly homologous to the Arabidopsis CYP707A4 protein (65% amino acid identity) and Solanum tuberosum CYP707A2 gene (95% identity). The tomato protein (designated SlCYP707A1) contains the highly conserved cysteine residue (within the PFGNGTHSCPG motif), which is the putative heme-iron-ligand, common to all P450s and essential for catalytic activity (Kushiro et al. 2004). A database search yielded three more putative CYP707A ESTs (TC177455, AI484420, and TC186477) that share between 61 and 68% sequence identity with SlCYP707A1 at nucleotide level. Their deduced protein sequences are each 70–74% identical with one of the four Arabidopsis AtCYP707A proteins. We, therefore, consider the three genes to encode putative ABA 8′-hydroxylases, and designated them SlCYP707A2, SlCYP707A3 and SlCYP707A4.
The mRNA levels of the four putative tomato CYP707A genes were determined in emasculated flowers 3 days after full bloom (control) and 3 days after pollination in tomato ovaries. Figure 3b shows that the mRNA concentration of SlCYP707A1 strongly increased after pollination in whole ovaries and it reached a much higher level than the mRNA levels of the other CYP707A genes. In additional, the mRNA levels of all other putative tomato CYP707A genes were reduced after pollination. This suggests that SlCYP707A1 is the most important gene responsible for ABA catabolism in pollinated ovaries. Expression of SlCYP707A1 was also analyzed in other tissues and it is expressed in sepals, petals and pedicels (Fig. 3c). The accumulation of SlCYP707A1 mRNA in ovaries was determined more precisely in separated ovule/placenta and pericarp tissue during the first 3 days after pollination. SlCYP707A1 mRNA levels were several hundredfold up-regulated after pollination, specifically in ovules/placenta and not in pericarp (Fig. 3d).
Furthermore, mRNA localization was analyzed in ovules and placenta separately. In unpollinated (control) ovaries SlCYP707A1 mRNA is present in the placenta, but not in the ovules and pericarp (Fig. 4a). In pollinated ovaries, SLCYP707A1 mRNA level is generally higher and present in both placenta and ovule tissue. In situ hybridization with a gene-specific probe confirmed this localization of SlCYP707A1 in pollinated ovaries. The purple staining suggests relatively high SlCYP707A1 mRNA levels in the ovules and in the periphery of the placenta but low levels in the pericarp (Fig. 4b, c).
Gene expression data suggest that the reduction in ABA levels that we measured after pollination could be due to increased ABA 8′-hydroxylation activity. To strengthen this hypothesis, we analyzed the levels of the ABA catabolic products in ovaries 3 days after pollination and 3 days after anthesis in unpollinated ovaries. These tissues are comparable to the C3 and 3 dap samples, respectively, which were used to measure ABA (Fig. 1). Table 2 shows that the level of ABA in unpollinated ovaries was two-and-half-fold higher than in pollinated ovaries, comparable to the measurements shown in Fig. 1. The product of ABA 8′-hydroxylation is (−)-PA which can be reduced to (−)-DPA (Zeevaart 1999). PA levels did not change, but DPA levels increased from 0.44 to 1.20 ng/mg DW in ovaries after pollination. 7′- and 9′-ABA hydroxylation can also inactivate ABA in several plant species (Nambara and Marion-Poll 2005). The product of ABA 9′-hydroxylation is 9′-hydroxy ABA which is converted to neo-phaseic acid (neo-PA). In tomato ovaries, a considerable amount of neo-PA was present, but this was not affected by pollination. The presence of 7′-hydroxy ABA was also detected, but its level was low and could not be quantified significantly. The amount of a conjugated form of ABA, ABA-glucosyl ester (ABA-GE), decreased from 1.32 to 0.65 ng/mg DW in ovaries after pollination (Table 2).
Functional analysis of SlCYP707A1
To prove that the SlCYP707A1 gene is coding for a functional ABA 8′-hydroxylase, we overexpressed its coding region under control of the 35S-promoter in tomato. Several transgenic lines were obtained, of which the line with the highest overexpression level was further analyzed. Figure 5a shows the SlCYP707A1 mRNA levels in unpollinated mature ovaries. The SlCYP707A1 overexpression line has approximately 45-fold higher mRNA levels than wild type. Consequently, the free-ABA level in unpollinated ovaries was 46% lower in SlCYP707A1 overexpression plants than in wild-type plants (Fig. 5b), confirming the ABA 8′-hydroxylase activity of SlCYP707A1. The overexpression plants have a smaller leaf surface area and wilt stronger 72 h after water with holding than wild-type plants (Fig. 5c, d). In addition, we also observed the initiation of adventitious root growth on the stem of the SlCYP707A1 overexpression plants (Fig. 5e).
a Relative mRNA levels of the SlCYP707A1 gene in unpollinated mature ovaries of overexpression (OE) plants are 45 times higher than wild type (wt). b Relative ABA levels in unpollinated mature ovaries of overexpression plants is 56% of wild type. c Overexpression plants (right) wilt stronger after 72 h of water with holding than wild-type plants (left). d The leaf surface area of the compound leaves of overexpression plants is smaller than wild type. e On the stem of the overexpression plants small adventitious roots are visible
Hormone crosstalk
Because fruit set is induced by GA and auxin, we were interested in the effect of these hormones on the activity of both, the LeNCED1 gene and the SlCYP707A1 gene. The LeNCED1 mRNA level in ovaries was lower after GA3 treatment or 4-Cl-IAA treatment and comparable to the levels found after pollination (Fig. 6a). ABA treatment, however, increased the mRNA level of LeNCED1. SlCYP707A1 mRNA level was not affected by GA3 treatment in contrast to pollination, 4-Cl-IAA treatment, or ABA treatment, which all had a stimulating effect (Fig. 6b).
mRNA levels of LeNCED1 and SlCYP707A1 before (C = control) and after hormone treatment or after pollination. a LeNCED1 expression is induced by ABA treatment and reduced after pollination, GA3 and 4-Cl-IAA (IAA) treatment. b SlCYP707A1 expression is induced after pollination, 4-Cl-IAA and ABA treatment. Mean values (n = 6) ± SE
Discussion
ABA content
Fruit set is an agro-economically important process, in which hormones play a pivotal role. Gibberellins and auxins were known previously to induce fruit growth and, by demonstrating that ABA signaling is high in mature ovaries and decreases after pollination, we have recently suggested a role for ABA in fruit set (Vriezen et al. 2008). In additional, ABA has been immuno-localized in the ovary of Arabidopsis and cucumber, indicating a function for ABA in these tissues (Peng et al. 2006). Previously, Kojima et al. (1993) had measured relatively high levels of ABA in unpollinated pistils compared to 10-day-old tomato fruits. Here, we demonstrate that within 3 days the free-ABA content of pollinated tomato ovaries decreases to approximately 30% of that in unpollinated ovaries. A decrease in ABA concentration to 50 or 25% of the original concentration has also been measured in other physiological systems, such as in dormancy breakage in seeds (Chiwocha et al. 2005) and tubers (Destefano-Beltran et al. 2006). The relative decrease in ABA content in the tomato ovary is comparable to those observed in other processes in which ABA level is of physiological importance.
Regulation of ABA biosynthesis and catabolism
Abscisic acid concentration is actively regulated in ovaries, as we found biosynthesis genes to be down-regulated and ABA catabolism genes to be induced in the ovary after fruit initiation. NCED genes are often encoded by a small gene family. However, we were not able to find putative NCED genes other than LeNCED1 in the tomato sequence databases, which contain 213,947 ESTs and 41,425 unique sequences. The three other CCDs described are probably not related to ABA biosynthesis as they do not group with LeNCED1. Moreover, TC154637 and TC154638 have already been described as carotenoid cleavage dioxygenase 1 genes involved in volatile terpenoid production (Simkin et al. 2004). Therefore, LeNCED1 appears to produce the most represented NCED transcript in ovary and possibly is the only NCED gene in tomato. In addition, the fact that the notabilis mutant with a mutation in the LeNCED1 gene has a strong ABA-deficient phenotype (Burbidge et al. 1999; Thompson et al. 2004) indicates that this gene has a prominent role in ABA biosynthesis. Transcript levels for this gene decreased after pollination concomitant with the decrease in ABA concentration, indicating that LeNCED1 regulation directly affects biosynthesis of ABA in the mature ovary.
Transcription levels of a gene very homologous to an Arabidopsis ABA 8′-hydroxylase (AtCYP707A4) were strongly induced specifically in ovules and placenta tissue, in contrast to the mRNA levels of the three other putative tomato CYP707A genes. Similarly, the CYP707A genes in Arabidopsis are also differentially expressed, for instance during dehydration and rehydration responses and during seed dormancy (Kushiro et al. 2004; Okamoto et al. 2006). The deduced protein sequence of SlCYP707A1 has very high homology to AtCYP707A4. Moreover, we have indirectly proven that the SlCYP707A1 gene encodes a functional ABA 8′-hydroxylase by overexpressing it in tomato. This resulted in reduced ABA levels and ABA-deficient phenotypes, such as adventitious rooting, reduced leaf surface area and increased wilting. These characteristics have also been described for the tomato ABA-deficient mutants sitiens, notabilis and flacca (Taylor and Tarr 1984; Burbidge et al. 1999). The remarkably large increase we found in SlCYP707A1 mRNA levels suggests a strong induction of ABA 8′-hydroxylation after pollination. In addition, we showed that after pollination a higher level of DPA, the end product of the ABA 8′-hydroxylation pathway, was present in the ovary, suggesting that ABA 8′-hydroxylation was indeed induced after pollination. Interestingly, 7′- and 9′-hydroxylation of ABA also takes place in tomato. However, no differences were found between 7′-hydroxy ABA and neo-PA levels in ovaries after pollination, suggesting that these conversions take place, but do not attribute to the decrease in ABA levels after pollination. Thus, ABA 8′-hydroxylation is the major ABA catabolic pathway in tomato ovaries (our work) as was suggested for Arabidopsis (Kushiro et al. 2004). ABA can be transported and stored as ABA-GE (Nambara and Marion-Poll 2005) and the high levels in unpollinated ovaries could mean that ABA-GE is transported to and stored in the ovary. Altogether, these data strengthen our hypothesis that after pollination ABA levels are reduced mainly through ABA 8′-hydroxylation. The specific localization of SlCYP707A1 in ovules and placenta might indicate that embryo or seed formation requires rapid removal of ABA. The localization of SlCYP707A1 mRNA is comparable to the localization of the hormone ABA in the ovules of Arabidopsis and cucumber (Peng et al. 2006). The decrease in mRNA levels of the three other putative tomato CYP707A genes might be explained by the reduction in ABA concentration, since positive feedback from ABA on CYP707A expression has been found in Arabidopsis (Kushiro et al. 2004; Umezawa et al. 2006), potato (Destefano-Beltran et al. 2006) and by us in tomato (Fig. 6b).
Hormone crosstalk
Auxin has been shown to interact with several hormones via regulation of expression of genes encoding biosynthetic or catabolic enzymes (Ross and O’Neill 2001). A fertilization-induced increase in auxin signaling most likely regulates the expression of ABA biosynthesis and catabolism genes during tomato fruit set, since we could show that auxin treatment reduced LeNCED1 transcript levels and increased SlCYP707A1 transcript levels. GA3 treatment also reduced LeNCED1 gene expression but it had no effect on SlCYP707A1 gene expression. Therefore, auxin, via GA, or both hormones together regulate LeNCED1 gene expression, whereas SlCYP707A1 expression seems to be regulated by auxin alone. This would be consistent with auxin inducing GA biosynthesis, but not vice versa (Vriezen et al. 2008; de Jong et al. 2009). It seems likely that a pollination-induced auxin increase precedes the GA increase, as summarized in Fig. 7. This hormonal signaling cascade may also lead to a reduction in the ABA content via changes in LeNCED1 and SlCYP707A1 expression. In the promoter of LeNCED1, auxin, GA, and ABA response elements were identified (Thompson et al. 2004), consistent with LeNCED1 being regulated by all three hormones. Since we found that LeNCED1 is positively regulated by ABA, a decrease of ABA content after pollination might also contribute to the decrease in its mRNA levels. A positive feedback of ABA levels on NCED mRNA levels has been shown in Arabidopsis (Wan and Li 2006). However, Thompson et al. (2000) showed that ABA had no effect on LeNCED1 expression in a detached leaf assay. Different tissues might thus have different regulatory mechanisms of LeNCED1 expression. The increase in SlCYP707A1 levels cannot be caused by a lower content of ABA, because this gene is also positively regulated by ABA.
Model of the hormonal interactions after pollination. Pollination induces an increase in auxin concentration and subsequently an increase in GA concentration. Both an increase in GA and IAA levels inhibit the expression of the LeNCED1 gene (ABA biosynthesis), while only increased IAA levels can stimulate the expression of the SlCYP707A1 gene (catabolism), ABA levels therefore decrease
Function of ABA during fruit set
In a mature unpollinated ovary, many cell cycle genes, such as LeCDKB1,2, CycA1,2 and CycD3, are relative lowly expressed (Vriezen et al. 2008). In addition, the relative expression level in the mature tomato ovary of two genes homologous to PsDRM1, which proved to be excellent dormancy (DRM) markers in pea (Stafstrom et al. 1998), are high. Together, this suggests that the mature ovary before pollination has entered a temporarily dormant state, during which the tissue is quiescent. It is known that ABA can directly influence the cell cycle via suppression of CDKA1 expression (Smalle et al. 2003) or stimulation of the ICK1 (an inhibitor of CDK action) expression (Wang et al. 1998), thereby inhibiting cell division and inducing an quiescent state. We know that the ABA signaling and response pathway are active in the unpollinated mature ovary (Vriezen et al. 2008). In addition, in the research presented here, we have shown that ABA levels are relatively high in mature ovaries and are down-regulated after pollination. Based on these observations, a hypothesis is that ABA inhibits growth until fruit set. The post-pollination increase of auxin and GA levels may directly or indirectly repress ABA biosynthesis. However, we were unable to induce fruit set by application of an ABA biosynthesis inhibitor, fluridone. Neither could we inhibit fruit set by ABA application to pollinated ovaries (data not shown). This might indicate that not only the concentration of ABA but rather the hormonal balance of ABA and other hormones, such as auxin and GA, is important. Similarly, the potential of axillary bud outgrowth is determined by a balance of the hormones such as cytokinin, auxin and ABA (Shimizu-Sato and Mori 2001). Besides inhibition of growth, ABA can also induce tolerance to several abiotic stresses, such as heat stress, cold stress and drought stress (Bartels and Sunkar 2005; Gusta et al. 2005; Bonham-Smith et al. 2008). For example, ABA induces dehydrins which are proteins with a potential in vivo role in stabilizing cells under abiotic stress (Kalemba and Pukacka 2007). Several dehydrins are indeed higher expressed in the mature unpollinated ovary (Vriezen et al. 2008). A putative protective function for ABA might become more apparent during unfavorable environmental conditions when ABA protects the mature ovary during its temporal dormant state.
Abbreviations
- ABA:
-
Abscisic acid
- GA:
-
Gibberellin
- NCED:
-
9-cis-Epoxy-carotenoid dioxygenase
- DPA:
-
Dihydrophaseic acid
- ABA-GE:
-
ABA-glucose ester
- PA:
-
Phaseic acid
- neo-PA:
-
neo-Phaseic acid
References
Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24:23–58
Bereterbide A, Hernould M, Farbos I, Glimelius K, Mouras A (2002) Restoration of stamen development and production of functional pollen in an alloplasmic CMS tobacco line by ectopic expression of the Arabidopsis thaliana SUPERMAN gene. Plant J 29:607–615
Bonham-Smith PC, Kapoor M, Bewley JD (2008) Exogenous application of abscisic acid or triadimefon affects the recovery of Zea mays seedlings from heat shock. Physiol Plant 73:27–30
Burbidge A, Grieve TM, Jackson A, Thompson A, McCarty DR, Taylor IB (1999) Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J 17:427–431
Chalfun-Junior A, Franken J, Mes JJ, Marsch-Martinez N, Pereira A, Angenent GC (2005) ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal–distal patterning in Arabidopsis petals. Plant Mol Biol 57:559–575
Chiwocha SDS, Cutler AJ, Abrams SR, Ambrose SJ, Yang J, Ross ARS, Kermode AR (2005) The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J 42:35–48
de Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH (2009) The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57:160–170
Destefano-Beltran L, Knauber D, Huckle L, Suttle JC (2006) Effects of postharvest storage and dormancy status on ABA content, metabolism, and expression of genes involved in ABA biosynthesis and metabolism in potato tuber tissues. Plant Mol Biol 61:687–697
Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59:387–415
Fos M, Nuez F, Garcia-Martinez JL (2000) The gene pat-2, which induces natural parthenocarpy, alters the gibberellin content in unpollinated tomato ovaries. Plant Physiol 122:471–479
Fos M, Proano K, Nuez F, Garcia-Martinez JL (2001) Role of gibberellins in parthenocarpic fruit development induced by the genetic system pat-3/pat-4 in tomato. Physiol Plant 111:545–550
Garcia-Martinez JL, Carbonell J (1980) Fruit-set of unpollinated ovaries of Pisum sativum L. influence of plant-growth regulators. Planta 147:451–456
Gillaspy G, Ben-David H, Gruissem W (1993) Fruits—a developmental perspective. Plant Cell 5:1439–1451
Goetz M, Hooper LC, Johnson SD, Macedo Rodrigues JC, Koltunow AM (2007) Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol 145:351–366
Gorguet B, van Heusden AW, Lindhout P (2005) Parthenocarpic fruit development in tomato. Plant Biol 7:131–139
Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, Thomas SG (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 12:3399–3414
Gusta LV, Trischuk R, Weiser CJ (2005) Plant cold acclimation: the role of abscisic acid. J Plant Growth Regul 24:308–318
Horvath DP, Anderson JV, Chao WS, Foley ME (2003) Knowing when to grow: signals regulating bud dormancy. Trends Plant Sci 8:534–540
Kalemba EM, Pukacka S (2007) Possible roles of LEA proteins and sHSPs in seed protection: a short review. Biol Lett 44:3–16
Kojima K, Kuraishi S, Sakurai N, Fusao K (1993) Distribution of abscisic-acid in different parts of the reproductive-organs of tomato. Sci Hort 56:23–30
Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23:1647–1656
Marti C, Orzaez D, Ellul P, Moreno V, Carbonell J, Granell A (2007) Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J 52:865–876
Mazzucato A, Taddei AR, Soressi GP (1998) The parthenocarpic fruit (pat) mutant of tomato (Lycopersicon esculentum Mill.) sets seedless fruits and has aberrant anther and ovule development. Development 125:107–114
Mezzetti B, Landi L, Pandolfini T, Spena A (2004) The defH9-iaaM auxin-synthesizing gene increases plant fecundity and fruit production in strawberry and raspberry. BMC Biotechnol 4:4–13
Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56:165–185
Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E (2006) CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 141:97–107
Olimpieri I, Siligato F, Caccia R, Mariotti L, Ceccarelli N, Soressi GP, Mazzucato A (2007) Tomato fruit set driven by pollination or by the parthenocarpic fruit allele are mediated by transcriptionally regulated gibberellin biosynthesis. Planta 226:877–888
Owen SJ, Abrams SR (2009) Measurement of plant hormones by liquid chromatography–mass spectrometry. In: Cutler S, Bonetta D (eds) Plant hormones: methods and protocols. Humana Press (in press)
Peng YB, Cheng Z, Wang DH, Gong HQ, Xu ZH, Bai SN (2006) Preferential localization of abscisic acid in primordial and nursing cells of reproductive organs of Arabidopsis and cucumber. New Phytol 170:459–466
Qin XQ, Zeevaart JAD (2002) Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol 128:544–551
Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang YY, Imai R, Sekimoto H, Kamiya Y (1999) Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. Plant J 17:241–250
Rodrigo MJ, Garcia-Martinez JL (1998) Hormonal control of parthenocarpic ovary growth by the apical shoot in pea. Plant Physiol 116:511–518
Ross J, O’Neill D (2001) New interactions between classical plant hormones. Trends Plant Sci 6:2–4
Rotino GL, Perri E, Zottini M, Sommer H, Spena A (1997) Genetic engineering of parthenocarpic plants. Nat Biotechnol 15:1398–1401
Serrani JC, Sanjuan R, Ruiz-Rivero O, Fos M, Garcia-Martinez JL (2007) Gibberellin regulation of fruit set and growth in tomato. Plant Physiol 145:246–257
Shimizu-Sato S, Mori H (2001) Control of outgrowth and dormancy in axillary buds. Plant Physiol 127:1405–1413
Simkin AJ, Schwartz SH, Auldridge M, Taylor MG, Klee H (2004) The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β-ionone pseudoionone, and geranylacetone. Plant J 40:882–892
Smalle J, Kurepa J, Yang PZ, Emborg TJ, Babiychuk E, Kushnir S, Vierstra RD (2003) The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15:965–980
Stafstrom JP, Ripley BD, Devitt ML, Drake B (1998) Dormancy-associated gene expression in pea axillary buds. Planta 205:547–552
Tan BC, Joseph LM, Deng WT, Liu LJ, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis-epoxycarotenoid dioxygenase gene family. Plant J 35:44–56
Taylor IB, Tarr AR (1984) Phenotypic interactions between abscisic acid deficient tomato mutants. Theor Appl Gen 68:115–119
Thompson AJ, Jackson AC, Parker RA, Morpeth DR, Burbidge A, Taylor IB (2000) Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol Biol 42:833–845
Thompson AJ, Thorne ET, Burbidge A, Jackson AC, Sharp RE, Taylor IB (2004) Complementation of notabilis, an abscisic acid-deficient mutant of tomato: importance of sequence context and utility of partial complementation. Plant Cell Environ 27:459–471
Umezawa T, Okamoto M, Kushiro T, Nambara E, Oono Y, Seki M, Kobayashi M, Koshiba T, Kamiya Y, Shinozaki K (2006) CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J 46:171–182
Vandesomple J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034.1–research0034.11
Vriezen WH, Feron R, Maretto F, Keijman J, Mariani C (2008) Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol 177:60–76
Wan XR, Li L (2006) Regulation of ABA level and water-stress tolerance of Arabidopsis by ectopic expression of a peanut 9-cis-epoxycarotenoid dioxygenase gene. Biochem Biophys Res Commun 347:1030–1038
Wang H, Qi QG, Schorr P, Cutler AJ, Crosby WL, Fowke LC (1998) ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J 15:501–510
Wang H, Jones B, Li ZG, Frasse P, Delalande C, Regad F, Chaabouni S, Latche A, Pech JC, Bouzayen M (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17:2676–2692
Zeevaart JAD (1999) Abscisic acid metabolism and its regulation. In: Hooykaas MA, Hall MA, Libbenga KR (eds) Biochemistry and molecular biology of plant hormones. Elsevier, New York, pp 189–207
Acknowledgments
We thank Dr. Ivo Rieu (University of Freiburg, Germany) for his cooperation and fruitful scientific discussions. Part of the research was funded by a grant from the Netherlands Organization for Scientific Research (NWO, R 89-182).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Nitsch, L.M.C., Oplaat, C., Feron, R. et al. Abscisic acid levels in tomato ovaries are regulated by LeNCED1 and SlCYP707A1 . Planta 229, 1335–1346 (2009). https://doi.org/10.1007/s00425-009-0913-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-009-0913-7