Abstract
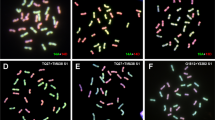
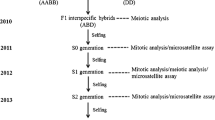
We used a newly synthesized allotetraploid between C. sativus (2n = 2x = 14, n gametic chromosome number, x haploid chromosome number) and C. hystrix (2n = 2x = 24) to study the genomic events in its early generations. Results from cytological characterization of the F1 and the allotetraploid progenies showed that the rate of bivalents in meiotic metaphase I of the F1 was greatly improved by chromosome doubling, and further improved during the selfing process of allopolyploid resulting into relatively diploid-like meiosis. Extensive genomic changes were detected by amplified fragment length polymorphism analysis. The changes mainly involved loss of parental restriction fragments and gaining of novel fragments. The total detectable changes were from 11.1 to 32.1%, and the frequency of losing parental fragments was much higher than that of gaining novel fragments. Some of the changes were initiated as early as in the F1 hybrid, whereas others occurred after chromosome doubling (polyploid formation). No significant differences were detected in the reciprocal F1 hybrids and S0 generations. But the data showed that the frequency of sequence losing in C. sativus was about two times higher than in the C. hystrix. Our results demonstrated that the sequence elimination was the major event of genomic changes, and it might provide the physical basis for the diploid-like meiotic behavior in the diploidization of the newly formed allopolyploids. Moreover, the results suggest that the sequence elimination was not caused by cytoplasmic factors, and might relate to genomic recombination and to the numbers of parental chromosome.



Similar content being viewed by others
Abbreviations
- AFLP:
-
Amplified fragment length polymorphism
- A I:
-
Anaphase I
- CTAB:
-
Cetyltrimethylammonium bromide
- M I:
-
Metaphase I
- MSAP:
-
Methylation-sensitive amplification polymorphism
- PCR:
-
Polymerase chain reaction
- PMC:
-
Pollen mother cell
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Axesson T, Bowman CM, Sharpe AG, Lydiate DJ, Lagercrantz U (2000) Amphidiploid Brassica juncea contains conserved progenitor genomes. Genome 43:679–688
Baumel A, Ainouche M, Kalendar R, Schulman AH (2002) Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C. E. Hubbard (Poaceae). Mol Biol Evol 19:1218–1227
Bennett ST, Kenton AY, Bennett MD (1992) Genomic in situ hybridization reveals the allopolyploid nature of Millium mantianum (Gramineae). Chromosoma 101:420–424
Chen JF, Kirkbride JJr (2000) A new synthetic species of Cucumis (Cucurbitaceae) from interspecific hybridization and chromosome doubling. Brittonia 52:315–319
Chen JF, Isshiki S, Tashiro Y, Miyazaki S (1995) Studies on a wild cucumber from China (Cucumis hystrix Chakr.).I.Genetic distances C. hysrtix and two cultivated Cucumis species(C. sativus L. and C. melo L.) based on isozyme analysis. J Jpn Soc Hort Sci 64(suppl. 2):264–265
Chen JF, Staub JE, Tashiro Y, Isshiki S, Miyazaki S (1997) Successful interspecific hybridization between Cucumis sativus L. and Cucumis hystrix Chakr. Euphytica 96:413–419
Chen JF, Staub JE, Jiang JM (1998) A reevaluation of karyotype in cucumber (Cucumis sativus L.). Genet Resour Crop Evol 45:301–305
Chen JF, Staub JE, Adelberg J, Lewis S, Kunlle B (2002) Synthesis and preliminary characterization of a new species (Amphidiploid) in Cucumis. Euphytica 123:315–322
Chen JF, Staub J, Qian CH, Luo XD, Zhuang FY (2003) Reproduction and cytogenetic characterization of interspecific hybrids from Cucumis hystix chakr. × C. sativus L. Theor Appl Genet 106:688–695
Chen JF, Luo XD, Qian CH, Molly MJ, Staub J, Zhuang FY, Lou QF, Ren G (2004) Cucumis monosomic alien addition lines: morphological, cytological, and genotypic analyses. Theor Appl Genet 108:1343–1348
Comai L (2000) Genetic and epigenetic interactions in allopolyploidy plants. Plant Mol Biol 43:387–399
Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y. Byer B (2000) Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allopolyploids. Plant Cell 12:1551–1567
Comai L, Tyagi AP, Lysak MA (2003) FISH analysis of meiosis in Arabidopsis allopolyploids. Chromosome Res 11:217–226
Contento A, Heslop-Harrison JS, Schwarzacher T (2005) Diversity of a major repetitive DNA sequence in diploid and polyploid Triticeae. Cytogenet Genome Res 109:34–42
Dvorak J, Zhang HB (1990) Variation in repeated nucleotide sequence sheds light on the phylogeny of the wheat B and G genome. Proc Natl Acad Sci USA 87:9640–9644
Eckert KA, Mowery A, Hile SE (2002) Misalignment-mediated DNA polymerase beta mutations: comparison of microsatellite frame-shift error rates using a forward mutation assay. Biochemistry 41:10490–10498
Feldman M, Levy AA (2005) Allopolyploidy–a shaping force in the evolution of wheat genomes. Cytogenet Genome Res 109:250–258
Feldman M, Liu B, Sehgal G, Abbo S, Levy AA, Vega JM (1997) Rapid elimination of low copy DNA sequence in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147:1381–1387
Helentjaris T, Weber D, Wright S (1988) Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics 118:353–363
Kirkbride J Jr (1993) Biosystematic monograph of the genus Cucumis (Cucurbitaceae). Parkway Publishers, Boone, pp 84–88
Lee HS, Chen ZJ (2000) Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc Natl Acad Sci USA 98:6753–6758
Leitch IJ, Bennett MD (1997) Polyploidy in angiosperms. Trends Plant Sci 2:470–476
Levin DA (1983) Polyploidy and novelty in flowering plants. Am Nat 122:1–25
Liu B, Wendel JF (2002) Non-mendelian phenomena in allopolyploid genome evolution. Curr Genomics 3:489–505
Liu B, Segal G, Vega JM (1997) Isolation and characterization of chromosome-specific DNA sequences from a chromosome arm genomic library of common wheat. Plant J 11:959
Liu B, Vega MJ, Feldman M (1998a) Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops II. Changes in low-copy coding DNA sequences. Genome 41:535–542
Liu B, Vega MJ, Segal G, Abbo S, Rodova M, Feldman M (1998b) Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. I. Changes in low-copy noncoding DNA sequences. Genome 41:272–277
Liu B, Brubaker CL, Mergeai G, Cronn RC, Wendel JF (2001) Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome 44:321–330
Lukens LN, Pires JC, Leon E, Vogelzang R, Oslach L, Osborn T (2006) Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol 140:336–348
Ma XF, Gustafson JP (2005) Genome evolution of allopolyploids: a process of cytological and genetic diploidization. Cytogenet Genome Res 109:1–3
Madlung A, Masuelli RW, Watson B, Reynolds SH, Davison J, Comai L (2002) Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allopolyploids. Plant Physiol 129:733–746
Masterson J (1994) Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264:421–424
McGire PE, Dvorak J (1982) Genetic regulation of heterogenetic chromosome paring in polyploid species of the genus Triticum Sensu lato. Can J Genet Cytol 24:57–82
Moore G (2002) Meiosis in allopolyploids the importance of “Teflon” chromosomes. Trends Genet 18:456–463
Murray HG, Thompson WF (1980) Rapid isolation of higher weight DNA. Nucleic Acids Res 8:4321
Ozkan H (2000) Genomic changes in newly synthesized amphiploids of Aegilops and Triticum. Ph.D. Thesis University of Cukurova
Ozkan H, Levy A, Feldman M (2001) Allopolyploid-induced rapid genomic evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13:1735–1747
Panaud O, Chen X, McCouch SD (1996) Development of microsatellite markers and characterization of sample sequence lengthen polymorphism (SSLP) in rice (Oryza sativa L.). Mol Gen Genet 252:597–607
Richard GF, Paques F (2000) Mini- and microsatellite expansions: the recombination connection. EMBO Rep 1:122–126
Shaked H, Kashkush K, Ozakan H, Feldman M, Levy AA (2001) Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13:1749–1759
Soltis DE, Soltis PS (1995) The dynamic nature of polyploid genome. Proc Natl Acad Sci USA 92:8089–8091
Song K, Lu P, Tang K, Osborn TC (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci USA 92:7719–7723
Stebbins GL (1971) Chromosomal evolution in higher plants. Addison-Wesley, New York
Thomas HM, Harper JA, Meredith MR, Morgan WG, King IP (1997) Physical mapping of ribosomal DNA sites in Festuca arundinacea and related species by in situ hybridization. Genome 40:406–410
Volkov RA, Borisjuk NV, Panchuk II, Schweizer D, Hem-leben V (1999) Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol Biol Evol 16:311–320
Vos P, Hogers R, Bleeker M, Reljans M, Lee T, Homes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249
Whitkus R, Doebley J, Lee M (1992) Comparative genome mapping of sorghum and maize. Genetics 132:1119–1130
Zohary D, Feldman M (1962) Hybridization between amphidiploids and the evolution of polyploids in the wheat (Aegilops-Triticum) group. Evolution 16:44–61
Acknowledgments
This research was partially supported by the General Program 30470120 from the National Natural Science Foundation of China, by the 863 Program 2004AA241120 from the Ministry of Science and Technology of China, and by the Ph.D Funding from the Ministry of Education of China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, L., Lou, Q., Zhuang, Y. et al. Cytological diploidization and rapid genome changes of the newly synthesized allotetraploids Cucumis × hytivus . Planta 225, 603–614 (2007). https://doi.org/10.1007/s00425-006-0381-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-006-0381-2