Abstract
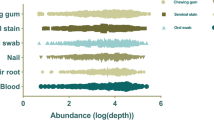
Short tandem repeats (STRs) are the preferred genetic markers in forensic DNA analysis, routinely measured by capillary electrophoresis (CE) method based on the fragment length features. While, the massive parallel sequencing (MPS) technology could simultaneously target a large number of intriguing forensic STRs, bypassing the intrinsic limitations of amplicon size separation and accessible fluorophores in CE, which is efficient and promising for enabling the identification of forensic biological evidence. Here, we developed a novel MPS-based Forensic Analysis System Multiplecues SetB Kit of 133-plex forensic STR markers (52 STRs and 81 Y-STRs) and one Y-InDel (M175) based on multiplex PCR and single-end 400 bp sequencing strategy. This panel was subjected to developmental validation studies according to the SWGDAM Validation Guidelines. Approximately 2185 MPS-based reactions using 6 human DNA standards and 8 male donors were conducted for substrate studies (filter paper, gauze, cotton swab, four different types of FTA cards, peripheral venous blood, saliva, and exfoliated cells), sensitivity studies (from 2 ng down to 0.0625 ng), mixture studies (two-person DNA mixtures), PCR inhibitor studies (seven commonly encountered PCR inhibitors), species specificity studies (11 non-human species), and repeatability studies. Results of concordance studies (413 Han males and 6 human DNA standards) generated by STRait Razor and in-house Python scripts indicated 99.98% concordance rate in STR calling relative to CE for STRs between 41,900 genotypes at 100 STR markers. Moreover, the limitations of present studies, the nomenclature rules and forensic MPS applications were also described. In conclusion, the validation studies based on ~ 2200 MPS-based and ~ 2500 CE-based DNA profiles demonstrated that the novel MPS-based panel meets forensic DNA quality assurance guidelines with robust, reliable, and reproducible performance on samples of various quantities and qualities, and the STR nomenclature rules should be further regulated to integrate the inconformity between MPS-based and CE-based methods.
Highlights
-
A novel MPS-based panel of 133-plex forensic STR markers (52 STRs and 81 Y-STRs) and one Y-InDel (M175) was developed based on multiplex PCR and single-end 400 bp sequencing strategy.
-
A reasonably large number of ~2200 MPS-based and ~2500 CE-based DNA profiles were generated to comprehensively validated the novel 133-plex forensic STR panel according to SWGDAM.
-
The concordance rate reached to 99.98% between 41900 MPS-based and CE-based genotypes at 100 STR markers (52 STRs and 48 Y-STRs).






Similar content being viewed by others
Data availability
The raw data of this article is available upon reasonable requests to the corresponding authors.
Code availability
The in-house Python scripts for length-based and sequence-based allele frequencies are available from the corresponding authors.
References
Gettings KB, Aponte RA, Vallone PM, Butler JM (2015) STR allele sequence variation: current knowledge and future issues. Forensic Sci Int Genet 18:118–130. https://doi.org/10.1016/j.fsigen.2015.06.005
Fregeau CJ, Fourney RM (1993) DNA typing with fluorescently tagged short tandem repeats: a sensitive and accurate approach to human identification. Biotechniques 15:100–119
Ziegle JS, Su Y, Corcoran KP et al (1992) Application of automated DNA sizing technology for genotyping microsatellite loci. Genomics 14:1026–1031. https://doi.org/10.1016/s0888-7543(05)80126-0
Butler JM (2015) The future of forensic DNA analysis. Philosophical transactions of the Royal Society of London Series B, Biological sciences 370. https://doi.org/10.1098/rstb.2014.0252
Young JM, Linacre A (2021) Massively parallel sequencing is unlocking the potential of environmental trace evidence. Forensic Sci Int Genet 50:102393. https://doi.org/10.1016/j.fsigen.2020.102393
Mori C, Matsumura S (2021) Current issues for mammalian species identification in forensic science: a review. Int J Legal Med 135:3–12. https://doi.org/10.1007/s00414-020-02341-w
Yu Z, Xie Q, Zhao Y, Duan L, Qiu P, Fan H (2021) NGS plus bacterial culture: a more accurate method for diagnosing forensic-related nosocomial infections. Leg Med 52:101910. https://doi.org/10.1016/j.legalmed.2021.101910
Young JM, Power D, Kanokwongnuwut P, Linacre A (2021) Ancestry and phenotype predictions from touch DNA using massively parallel sequencing. Int J Legal Med 135:81–89. https://doi.org/10.1007/s00414-020-02398-7
Haas C, Neubauer J, Salzmann AP, Hanson E, Ballantyne J (2021) Forensic transcriptome analysis using massively parallel sequencing. Forensic Sci Int Genet 52:102486. https://doi.org/10.1016/j.fsigen.2021.102486
Adolfsson E, Qvick A, Green H et al (2021) Technical in-depth comparison of two massive parallel DNA-sequencing methods for formalin-fixed paraffin-embedded tissue from victims of sudden cardiac death. Forensic Sci Int Genet 53:102522. https://doi.org/10.1016/j.fsigen.2021.102522
Cartozzo C, Simmons T, Swall J, Singh B (2021) Postmortem submersion interval (PMSI) estimation from the microbiome of Sus scrofa bone in a freshwater river. Forensic Sci Int 318:110480. https://doi.org/10.1016/j.forsciint.2020.110480
Lewis EJ, Weaver E, Hoyle A, Lagace R, Oldoni F, Podini D (2021) Retrofitting massively parallel sequencing (MPS) for HLA-DQA1 and polymarker (PM) in forensic casework. Int J Legal Med. https://doi.org/10.1007/s00414-021-02647-3
Linacre A (2021) Animal forensic genetics. Genes 12. https://doi.org/10.3390/genes12040515
He G, Liu J, Wang M et al (2021) Massively parallel sequencing of 165 ancestry-informative SNPs and forensic biogeographical ancestry inference in three southern Chinese Sinitic/Tai-Kadai populations. Forensic Sci Int Genet 52:102475. https://doi.org/10.1016/j.fsigen.2021.102475
Truelsen D, Pereira V, Phillips C, Morling N, Borsting C (2021) Evaluation of a custom GeneRead massively parallel sequencing assay with 210 ancestry informative SNPs using the Ion S5 and MiSeq platforms. Forensic Sci Int Genet 50:102411. https://doi.org/10.1016/j.fsigen.2020.102411
Chen QF, Kang KL, Song JJ et al (2021) Allelic diversity and forensic estimations of the Beijing Hans: comparative data on sequence-based and length-based STRs. Forensic Sci Int Genet 51:102424. https://doi.org/10.1016/j.fsigen.2020.102424
Senovska A, Drozdova E, Vaculik O et al (2021) Cost-effective straightforward method for captured whole mitogenome sequencing of ancient DNA. Forensic Sci Int 319:110638. https://doi.org/10.1016/j.forsciint.2020.110638
Beasley J, Shorrock G, Neumann R, May CA, Wetton JH (2021) Massively parallel sequencing and capillary electrophoresis of a novel panel of falcon STRs: concordance with minisatellite DNA profiles from historical wildlife crime. Forensic Sci Int Genet 54:102550. https://doi.org/10.1016/j.fsigen.2021.102550
Gorden EM, Sturk-Andreaggi K, Marshall C (2021) Capture enrichment and massively parallel sequencing for human identification. Forensic Sci Int Genet 53:102496. https://doi.org/10.1016/j.fsigen.2021.102496
Gross TE, Fleckhaus J, Schneider PM (2021) Progress in the implementation of massively parallel sequencing for forensic genetics: results of a European-wide survey among professional users. Int J Legal Med 135:1425–1432. https://doi.org/10.1007/s00414-021-02569-0
Ta MTA, Nguyen NN, Tran DM et al (2021) Massively parallel sequencing of human skeletal remains in Vietnam using the precision ID mtDNA control region panel on the Ion S5 system. Int J Legal Med. https://doi.org/10.1007/s00414-021-02649-1
Wang Q, Jin B, Liu F et al (2021) DNA-based eyelid trait prediction in Chinese Han population. Int J Legal Med. https://doi.org/10.1007/s00414-021-02570-7
Wu R, Li H, Li R et al (2021) Identification and sequencing of 59 highly polymorphic microhaplotypes for analysis of DNA mixtures. Int J Legal Med 135:1137–1149. https://doi.org/10.1007/s00414-020-02483-x
Phillips C (2017) A genomic audit of newly-adopted autosomal STRs for forensic identification. Forensic Sci Int Genet 29:193–204. https://doi.org/10.1016/j.fsigen.2017.04.011
Almalki N, Chow HY, Sharma V, Hart K, Siegel D, Wurmbach E (2017) Systematic assessment of the performance of illumina’s MiSeq FGx forensic genomics system. Electrophoresis 38:846–854. https://doi.org/10.1002/elps.201600511
de Knijff P (2019) From next generation sequencing to now generation sequencing in forensics. Forensic Sci Int Genet 38:175–180. https://doi.org/10.1016/j.fsigen.2018.10.017
Ballard D, Winkler-Galicki J, Wesoly J (2020) Massive parallel sequencing in forensics: advantages, issues, technicalities, and prospects. Int J Legal Med 134:1291–1303. https://doi.org/10.1007/s00414-020-02294-0
Fan H, Du Z, Wang F et al (2021) The forensic landscape and the population genetic analyses of Hainan Li based on massively parallel sequencing DNA profiling. Int J Legal Med 135:1295–1317. https://doi.org/10.1007/s00414-021-02590-3
Calafell F, Anglada R, Bonet N et al (2016) An assessment of a massively parallel sequencing approach for the identification of individuals from mass graves of the Spanish Civil War (1936–1939). Electrophoresis 37:2841–2847. https://doi.org/10.1002/elps.201600180
Cheng K, Skillman J, Hickey S et al (2020) Variability and additivity of read counts for aSTRs in NGS DNA profiles. Forensic Sci Int Genet 48:102351. https://doi.org/10.1016/j.fsigen.2020.102351
Churchill JD, Schmedes SE, King JL, Budowle B (2016) Evaluation of the Illumina((R)) Beta Version ForenSeq DNA Signature Prep Kit for use in genetic profiling. Forensic Sci Int Genet 20:20–29. https://doi.org/10.1016/j.fsigen.2015.09.009
Delest A, Godfrin D, Chantrel Y et al (2020) Sequenced-based French population data from 169 unrelated individuals with Verogen’s ForenSeq DNA signature prep kit. Forensic Sci Int Genet 47:102304. https://doi.org/10.1016/j.fsigen.2020.102304
England R, Nancollis G, Stacey J, Sarman A, Min J, Harbison S (2020) Compatibility of the ForenSeq DNA Signature Prep Kit with laser microdissected cells: an exploration of issues that arise with samples containing low cell numbers. Forensic Sci Int Genet 47:102278. https://doi.org/10.1016/j.fsigen.2020.102278
Fregeau CJ (2021) Validation of the Verogen ForenSeq DNA Signature Prep kit/Primer Mix B for phenotypic and biogeographical ancestry predictions using the Micro MiSeq(R) flow cells. Forensic Sci Int Genet 53:102533. https://doi.org/10.1016/j.fsigen.2021.102533
Guevara EK, Palo JU, King JL et al (2021) Autosomal STR and SNP characterization of populations from the Northeastern Peruvian Andes with the ForenSeq DNA Signature Prep Kit. Forensic Sci Int Genet 52:102487. https://doi.org/10.1016/j.fsigen.2021.102487
Guo F, Yu J, Zhang L, Li J (2017) Massively parallel sequencing of forensic STRs and SNPs using the Illumina((R)) ForenSeq DNA Signature Prep Kit on the MiSeq FGx Forensic Genomics System. Forensic Sci Int Genet 31:135–148. https://doi.org/10.1016/j.fsigen.2017.09.003
Hussing C, Bytyci R, Huber C, Morling N, Borsting C (2019) The Danish STR sequence database: duplicate typing of 363 Danes with the ForenSeq DNA Signature Prep Kit. Int J Legal Med 133:325–334. https://doi.org/10.1007/s00414-018-1854-0
Jager AC, Alvarez ML, Davis CP et al (2017) Developmental validation of the MiSeq FGx Forensic Genomics System for targeted next generation sequencing in forensic DNA casework and database laboratories. Forensic Sci Int Genet 28:52–70. https://doi.org/10.1016/j.fsigen.2017.01.011
Khubrani YM, Hallast P, Jobling MA, Wetton JH (2019) Massively parallel sequencing of autosomal STRs and identity-informative SNPs highlights consanguinity in Saudi Arabia. Forensic Sci Int Genet 43:102164. https://doi.org/10.1016/j.fsigen.2019.102164
Khubrani YM, Jobling MA, Wetton JH (2020) Massively parallel sequencing of sex-chromosomal STRs in Saudi Arabia reveals patrilineage-associated sequence variants. Forensic Sci Int Genet 49:102402. https://doi.org/10.1016/j.fsigen.2020.102402
Kocher S, Muller P, Berger B et al (2018) Inter-laboratory validation study of the ForenSeq DNA Signature Prep Kit. Forensic Sci Int Genet 36:77–85. https://doi.org/10.1016/j.fsigen.2018.05.007
Li H, Zhang C, Song G et al (2021) Concordance and characterization of massively parallel sequencing at 58 STRs in a Tibetan population. Mol Genet Genomic Med 9:e1626. https://doi.org/10.1002/mgg3.1626
Li R, Li H, Peng D et al (2019) Improved pairwise kinship analysis using massively parallel sequencing. Forensic Sci Int Genet 38:77–85. https://doi.org/10.1016/j.fsigen.2018.10.006
Muller P, Sell C, Hadrys T et al (2020) Inter-laboratory study on standardized MPS libraries: evaluation of performance, concordance, and sensitivity using mixtures and degraded DNA. Int J Legal Med 134:185–198. https://doi.org/10.1007/s00414-019-02201-2
Peng D, Zhang Y, Ren H et al (2020) Identification of sequence polymorphisms at 58 STRs and 94 iiSNPs in a Tibetan population using massively parallel sequencing. Sci Rep 10:12225. https://doi.org/10.1038/s41598-020-69137-1
Ren ZL, Zhang JR, Zhang XM et al (2021) Forensic nanopore sequencing of STRs and SNPs using Verogen’s ForenSeq DNA Signature Prep Kit and MinION. Int J Legal Med. https://doi.org/10.1007/s00414-021-02604-0
Sharma V, Jani K, Khosla P, Butler E, Siegel D, Wurmbach E (2019) Evaluation of ForenSeq Signature Prep Kit B on predicting eye and hair coloration as well as biogeographical ancestry by using Universal Analysis Software (UAS) and available web-tools. Electrophoresis 40:1353–1364. https://doi.org/10.1002/elps.201800344
Sharma V, van der Plaat DA, Liu Y, Wurmbach E (2020) Analyzing degraded DNA and challenging samples using the ForenSeq DNA Signature Prep kit. Sci Justice 60:243–252. https://doi.org/10.1016/j.scijus.2019.11.004
Shen X, Li R, Li H et al (2021) Noninvasive prenatal paternity testing with a combination of well-established SNP and STR markers using massively parallel sequencing. Genes 12. https://doi.org/10.3390/genes12030454
Szargut M, Diepenbroek M, Zielinska G et al (2019) Is MPS always the answer? Use of two PCR-based methods for Y-chromosomal haplotyping in highly and moderately degraded bone material. Forensic Sci Int Genet 42:181–189. https://doi.org/10.1016/j.fsigen.2019.07.016
Turrina S, De Leo D (2021) Resizing reaction volumes for the ForenSeq DNA Signature Prep kit library preparation. Med Sci Law 61:92–95. https://doi.org/10.1177/0025802420923163
Xavier C, Parson W (2017) Evaluation of the Illumina ForenSeq DNA Signature Prep Kit - MPS forensic application for the MiSeq FGx benchtop sequencer. Forensic Sci Int Genet 28:188–194. https://doi.org/10.1016/j.fsigen.2017.02.018
Xu M, Du Q, Ma G et al (2019) Utility of ForenSeq DNA Signature Prep Kit in the research of pairwise 2nd-degree kinship identification. Int J Legal Med 133:1641–1650. https://doi.org/10.1007/s00414-019-02003-6
Tao R, Wang S, Chen A et al (2021) Parallel sequencing of 87 STR and 294 SNP markers using the prototype of the SifaMPS panel on the MiSeq FGx system. Forensic Sci Int Genet 52:102490. https://doi.org/10.1016/j.fsigen.2021.102490
Li R, Shen X, Chen H, Peng D, Wu R, Sun H (2021) Developmental validation of the MGIEasy Signature Identification Library Prep Kit, an all-in-one multiplex system for forensic applications. Int J Legal Med 135:739–753. https://doi.org/10.1007/s00414-021-02507-0
Miao X, Shen Y, Gong X et al (2021) A novel forensic panel of 186-plex SNPs and 123-plex STR loci based on massively parallel sequencing. Int J Legal Med 135:709–718. https://doi.org/10.1007/s00414-020-02403-z
Barrio PA, Martin P, Alonso A et al (2019) Massively parallel sequence data of 31 autosomal STR loci from 496 Spanish individuals revealed concordance with CE-STR technology and enhanced discrimination power. Forensic Sci Int Genet 42:49–55. https://doi.org/10.1016/j.fsigen.2019.06.009
Ganschow S, Silvery J, Tiemann C (2019) Development of a multiplex forensic identity panel for massively parallel sequencing and its systematic optimization using design of experiments. Forensic Sci Int Genet 39:32–43. https://doi.org/10.1016/j.fsigen.2018.11.023
Liu Q, Ma G, Du Q et al (2020) Development of an NGS panel containing 42 autosomal STR loci and the evaluation focusing on secondary kinship analysis. Int J Legal Med 134:2005–2014. https://doi.org/10.1007/s00414-020-02295-z
Pang JB, Rao M, Chen QF et al (2020) A 124-plex Microhaplotype panel based on next-generation sequencing developed for forensic applications. Sci Rep 10:1945. https://doi.org/10.1038/s41598-020-58980-x
Silvery J, Ganschow S, Wiegand P, Tiemann C (2020) Developmental validation of the monSTR identity panel, a forensic STR multiplex assay for massively parallel sequencing. Forensic Sci Int Genet 46:102236. https://doi.org/10.1016/j.fsigen.2020.102236
Valle-Silva GD, Souza FDN, Marcorin L et al (2019) Applicability of the SNPforID 52-plex panel for human identification and ancestry evaluation in a Brazilian population sample by next-generation sequencing. Forensic Sci Int Genet 40:201–209. https://doi.org/10.1016/j.fsigen.2019.03.003
Wu L, Chu X, Zheng J et al (2019) Targeted capture and sequencing of 1245 SNPs for forensic applications. Forensic Sci Int Genet 42:227–234. https://doi.org/10.1016/j.fsigen.2019.07.006
Phillips C, Gettings KB, King JL et al (2018) “The devil’s in the detail”: release of an expanded, enhanced and dynamically revised forensic STR Sequence Guide. Forensic Sci Int Genet 34:162–169. https://doi.org/10.1016/j.fsigen.2018.02.017
Li W, Wang X, Wang X et al (2020) Forensic characteristics and phylogenetic analyses of one branch of Tai-Kadai language-speaking Hainan Hlai (Ha Hlai) via 23 autosomal STRs included in the Huaxia() Platinum System. Mol Genet Genomic Med 8:e1462. https://doi.org/10.1002/mgg3.1462
Wang F, Du Z, Han B et al (2021) Genetic diversity, forensic characteristics and phylogenetic analysis of the Qiongzhong aborigines residing in the tropical rainforests of Hainan Island via 19 autosomal STRs. Ann Hum Biol 48:335–342. https://doi.org/10.1080/03014460.2021.1951352
Fan H, Wang X, Ren Z et al (2019) Population data of 19 autosomal STR loci in the Li population from Hainan Province in southernmost China. Int J Legal Med 133:429–431. https://doi.org/10.1007/s00414-018-1828-2
Fan H, Wang X, Chen H, Li W, Wang W, Deng J (2019) The Ong Be language-speaking population in Hainan Island: genetic diversity, phylogenetic characteristics and reflections on ethnicity. Mol Biol Rep 46:4095–4103. https://doi.org/10.1007/s11033-019-04859-8
Fan H, Zeng Y, Wu W et al (2021) The Y-STR landscape of coastal southeastern Han: forensic characteristics, haplotype analyses, mutation rates, and population genetics. Electrophoresis 42:1578–1593. https://doi.org/10.1002/elps.202100037
Liu J, Wang R, Shi J et al (2020) The construction and application of a new 17-plex Y-STR system using universal fluorescent PCR. Int J Legal Med 134:2015–2027. https://doi.org/10.1007/s00414-020-02291-3
Fu J, Cheng J, Wei C, Khan MA, Jin Z, Fu J (2020) Assessing 23 Y-STR loci mutation rates in Chinese Han father-son pairs from southwestern China. Mol Biol Rep 47:7755–7760. https://doi.org/10.1007/s11033-020-05851-3
Lin H, Ye Q, Tang P, Mo T, Yu X, Tang J (2020) Analyzing genetic polymorphism and mutation of 44 Y-STRs in a Chinese Han population of Southern China. Leg Med 42:101643. https://doi.org/10.1016/j.legalmed.2019.101643
Mo XT, Zhang J, Ma WH et al (2019) Developmental validation of the DNATyper()Y26 PCR amplification kit: an enhanced Y-STR multiplex for familial searching. Forensic Sci Int Genet 38:113–120. https://doi.org/10.1016/j.fsigen.2018.10.008
Wang Q, Jin B, An G et al (2019) Rapidly mutating Y-STRs study in Chinese Yi population. Int J Legal Med 133:45–50. https://doi.org/10.1007/s00414-018-1894-5
Wu W, Ren W, Hao H et al (2018) Mutation rates at 42 Y chromosomal short tandem repeats in Chinese Han population in Eastern China. Int J Legal Med 132:1317–1319. https://doi.org/10.1007/s00414-018-1784-x
Yang Y, Wang W, Cheng F et al (2018) Haplotypic polymorphisms and mutation rate estimates of 22 Y-chromosome STRs in the Northern Chinese Han father-son pairs. Sci Rep 8:7135. https://doi.org/10.1038/s41598-018-25362-3
Fan H, Wang X, Chen H et al (2018) The evaluation of forensic characteristics and the phylogenetic analysis of the Ong Be language-speaking population based on Y-STR. Forensic Sci Int Genet 37:e6–e11. https://doi.org/10.1016/j.fsigen.2018.09.008
Luo C, Duan L, Li Y et al (2021) Insights from Y-STRs: forensic characteristics, genetic affinities, and linguistic classifications of Guangdong Hakka and She groups. Front Genet 12:676917. https://doi.org/10.3389/fgene.2021.676917
Fan H, Xie Q, Li Y, Wang L, Wen SQ, Qiu P (2021) Insights into forensic features and genetic structures of Guangdong Maoming Han bsased on 27 Y-STRs. Front Genet 12:690504. https://doi.org/10.3389/fgene.2021.690504
Fan H, Wang X, Chen H et al (2018) Population analysis of 27 Y-chromosomal STRs in the Li ethnic minority from Hainan province, southernmost China. Forensic Sci Int Genet 34:e20–e22. https://doi.org/10.1016/j.fsigen.2018.01.007
Fan H, Zhang X, Wang X et al (2018) Genetic analysis of 27 Y-STR loci in Han population from Hainan province, southernmost China. Forensic Sci Int Genet 33:e9–e10. https://doi.org/10.1016/j.fsigen.2017.12.009
Ding J, Fan H, Zhou Y et al (2020) Genetic polymorphisms and phylogenetic analyses of the Ü-Tsang Tibetan from Lhasa based on 30 slowly and moderately mutated Y-STR loci. Forensic Sci Res: 1-8. https://doi.org/10.1080/20961790.2020.1810882
Goodwin S, McPherson JD, McCombie WR (2016) Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17:333–351. https://doi.org/10.1038/nrg.2016.49
Korostin D, Kulemin N, Naumov V, Belova V, Kwon D, Gorbachev A (2020) Comparative analysis of novel MGISEQ-2000 sequencing platform vs Illumina HiSeq 2500 for whole-genome sequencing. PLoS ONE 15:e0230301. https://doi.org/10.1371/journal.pone.0230301
Woerner AE, King JL, Budowle B (2017) Fast STR allele identification with STRait Razor 3.0. Forensic Sci Int Genet 30:18–23. https://doi.org/10.1016/j.fsigen.2017.05.008
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
McKenna A, Hanna M, Banks E et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Van der Auwera GA, Carneiro MO, Hartl C et al (2013) From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Current protocols in bioinformatics 43: 11 0 1- 0 33. https://doi.org/10.1002/0471250953.bi1110s43
Bieber FR, Buckleton JS, Budowle B, Butler JM, Coble MD (2016) Evaluation of forensic DNA mixture evidence: protocol for evaluation, interpretation, and statistical calculations using the combined probability of inclusion. BMC Genet 17:125. https://doi.org/10.1186/s12863-016-0429-7
Alsafiah HM, Aljanabi AA, Hadi S, Alturayeif SS, Goodwin W (2019) An evaluation of the SureID 23comp Human Identification Kit for kinship testing. Sci Rep 9:16859. https://doi.org/10.1038/s41598-019-52838-7
Bai X, Yao Y, Wang C et al (2019) Development of a new 25plex STRs typing system for forensic application. Electrophoresis 40:1662–1676. https://doi.org/10.1002/elps.201900021
Li J, Luo H, Song F et al (2017) Validation of the Microreader 23sp ID system: a new STR 23-plex system for forensic application. Forensic Sci Int Genet 27:67–73. https://doi.org/10.1016/j.fsigen.2016.12.005
Peng H, Li X, Yao T et al (2020) Forensic effectiveness and genetic distribution of 20 non-CODIS STRs and D3S1358 in Guangdong Han population. Int J Legal Med 134:1319–1321. https://doi.org/10.1007/s00414-019-02094-1
Zhang J, Zhang J, Tao R et al (2020) A newly devised multiplex assay of novel polymorphic non-CODIS STRs as a valuable tool for forensic application. Forensic Sci Int Genet 48:102341. https://doi.org/10.1016/j.fsigen.2020.102341
Zhou Z, Shao C, Xie J et al (2020) Genetic polymorphism and phylogenetic analyses of 21 non-CODIS STR loci in a Chinese Han population from Shanghai. Mol Genet Genomic Med 8:e1083. https://doi.org/10.1002/mgg3.1083
Phillips C, Parson W, Amigo J et al (2016) D5S2500 is an ambiguously characterized STR: identification and description of forensic microsatellites in the genomics age. Forensic Sci Int Genet 23:19–24. https://doi.org/10.1016/j.fsigen.2016.03.002
Acknowledgements
First, we would like to thank all donors for this study. In addition, especially, we would like to thank DeepReads Biotech, Guangzhou, Guangdong, China for their technical assistance in support of this work.
Funding
This study was supported by the Program of Hainan Association for Science and Technology Plans to Youth R&D Innovation (QCXM201705), the Science and Technology Program of Guangzhou, China (No. 2019030016), the National Undergraduate Innovation and Entrepreneurship Training Program (No. 201911810008 and No. 201911810023), and the National Natural Science Foundation of China (NSFC, No. 32070576).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Fudan University of Life Sciences (No. 14012) and in accordance with the standards of the Declaration of Helsinki. All participants had signed the informed consent form.
Consent for publication
(Not applicable).
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fan, H., Wang, L., Liu, C. et al. Development and validation of a novel 133-plex forensic STR panel (52 STRs and 81 Y-STRs) using single-end 400 bp massive parallel sequencing. Int J Legal Med 136, 447–464 (2022). https://doi.org/10.1007/s00414-021-02738-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-021-02738-1