Abstract
The monophyletic duckweeds comprising five genera within the monocot order Alismatales are neotenic, free-floating, aquatic organisms with fast vegetative propagation. Some species are considered for efficient biomass production, for life stock feeding, and for (simultaneous) wastewater phytoremediation. The ancestral genus Spirodela consists of only two species, Spirodela polyrhiza and Spirodela intermedia, both with a similar small genome (~160 Mbp/1C). Reference genome drafts and a physical map of 96 BACs on the 20 chromosome pairs of S. polyrhiza strain 7498 are available and provide useful tools for further evolutionary studies within and between duckweed genera. Here we applied sequential comparative multicolor fluorescence in situ hybridization (mcFISH) to address homeologous chromosomes in S. intermedia (2n = 36), to detect chromosome rearrangements between both species and to elucidate the mechanisms which may have led to the chromosome number alteration after their evolutionary separation. Ten chromosome pairs proved to be conserved between S. polyrhiza and S. intermedia, the remaining ones experienced, depending on the assumed direction of evolution, translocations, inversion, and fissions, respectively. These results represent a first step to unravel karyotype evolution among duckweeds and are anchor points for future genome assembly of S. intermedia.








Similar content being viewed by others
References
Appenroth K-J, Teller S, Horn M (1996) Photophysiology of turion formation and germination in Spirodela polyrhiza. Biol Plant 38(1):95–106. doi:10.1007/bf02879642
Bog M, Lautenschlager U, Landrock MF, Landolt E, Fuchs J, Sowjanya Sree K, Oberprieler C, Appenroth K-J (2015) Genetic characterization and barcoding of taxa in the genera Landoltia and Spirodela (Lemnaceae) by three plastidic markers and amplified fragment length polymorphism (AFLP). Hydrobiologia 749(1):169–182. doi:10.1007/s10750-014-2163-3
Cao HX, Vu GT, Wang W, Appenroth KJ, Messing J, Schubert I (2016) The map-based genome sequence of Spirodela polyrhiza aligned with its chromosomes, a reference for karyotype evolution. New Phytol 209(1):354–363. doi:10.1111/nph.13592
Chamala S, Chanderbali AS, Der JP, Lan T, Walts B, Albert VA, dePamphilis CW, Leebens-Mack J, Rounsley S, Schuster SC et al (2013) Assembly and validation of the genome of the nonmodel basal angiosperm Amborella. Science 342(6165):1516–1517. doi:10.1126/science.1241130
Geber G (1989) Zur Karyosystematik der Lemnaceae. PhD thesis University of Vienna, Vienna, Austria.:140
Gottlob-McHugh SG, Levesque M, MacKenzie K, Olson M, Yarosh O, Johnson DA (1990) Organization of the 5S rRNA genes in the soybean glycine max (L.) Merrill and conservation of the 5S rDNA repeat structure in higher plants. Genome 33(4):486–494
Heslop-Harrison JS, Harrison GE, Leitch IJ (1992) Reprobing of DNA: DNA in situ hybridization preparations. Trends Genet 8(11):372–373
Ijdo JW, Wells RA, Baldini A, Reeders ST (1991) Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res 19(17):4780
Landolt E (1986) The family of Lemnaceae—a monographic study. Veröffentlichungen des Geobotanischen Institutes der Eidg Techn Hochschule, Zürich( 71):566 pp
Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci U S A 103(13):5224–5229. doi:10.1073/pnas.0510791103
Ma L, Vu GT, Schubert V, Watanabe K, Stein N, Houben A, Schubert I (2010) Synteny between Brachypodium distachyon and Hordeum vulgare as revealed by FISH. Chromosom Res 18(7):841–850. doi:10.1007/s10577-010-9166-3
Mandakova T, Lysak MA (2008) Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae). Plant Cell 20(10):2559–2570. doi:10.1105/tpc.108.062166
Mandakova T, Joly S, Krzywinski M, Mummenhoff K, Lysak MA (2010) Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell 22(7):2277–2290. doi:10.1105/tpc.110.074526
Michael TP, Bryant D, Gutierrez R, Borisjuk N, Chu P, Zhang H, Xia J, Zhou J, Peng H, El Baidouri M et al (2017) Comprehensive definition of genome features in Spirodela polyrhiza by high-depth physical mapping and short-read DNA sequencing strategies. Plant J. doi:10.1111/tpj.13400
Shibata F, Sahara K, Naito Y, Yasukochi Y (2009) Reprobing multicolor FISH preparations in lepidopteran chromosome. Zool Sci 26(3):187–190. doi:10.2108/zsj.26.187
Urbanska-Worytkiewicz K (1980) Cytological variation within the family of “Lemnaceae”. Veröffentlichungen des Geobotanischen Institutes der Eidg Tech Hochschule, Stiftung Rübel, in Zürich doi: 10.5169/seals-308615
Wang W, Kerstetter RA, Michael TP (2011) Evolution of genome size in duckweeds (Lemnaceae). J Bot 2011:1–9. doi:10.1155/2011/570319
Wang W, Haberer G, Gundlach H, Glasser C, Nussbaumer T, Luo MC, Lomsadze A, Borodovsky M, Kerstetter RA, Shanklin J et al (2014) The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat Commun 5:3311. doi:10.1038/ncomms4311
Acknowledgements
The authors thank Jörg Fuchs, IPK Gatersleben, Klaus Appenroth, Friedrich-Schiller-Universität, and Jena for discussions and critical reading of the manuscript and JF additionally for help with artwork. This study is supported by a grant of the German Research Foundation (SCHU 951/18-1) to IS. PNTH was supported by a PhD scholarship of the Vietnam Ministry of Education and Training.
Author information
Authors and Affiliations
Contributions
PNTH and IS designed experiments; PNTH performed experiments; PNTH and IS analyzed data; PNTH and IS wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplemental Figure 1
Location of pseudomolecule 21b in S. polyrhiza (PDF 98kb). a) BAC 037I18 (Ψ21b) does not co-localize with 004N06 (Ψ7) and 009L02 (Ψ32); (b) BACs 006D12 and 037I18 (Ψ21b) co-localized with BACs of Ψ16 (040G15, 036F14, 003B08) and Ψ30 (006A07). According to the chromosomal position of signals, Ψ21b (red and yellow signals) appears in terminal position on ChrSp14, (c) ChrS17 includes only Ψ7 and Ψ32 (red arrow), and ChrS14 includes Ψ21b, Ψ16 and Ψ30 (yellow arrow). Probes were labelled by Cy3 (yellow), Alexa-488 (green) and Texas-Red (red), the chromosomes were counterstained with DAPI. Scale bar = 5 µm
Supplemental Figure 2
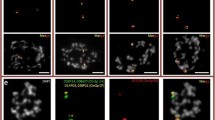
Chromosomal location of the 20 S. polyrhiza chromosome-specific probes on S. intermedia metaphases (PDF 240kb). (a) FISH signals of 20 S. polyrhiza chromosome-specific probes on two different S. intermedia metaphases confirmed 10 conserved chromosome pairs, the six new linkages (indicated by two-color arrow heads according to the false colors of signals), and parts of rearranged chromosomes that stay separately in S. intermedia, (b) Positions of 20 S. polyrhiza chromosome-specific probes are summarized: In the upper cell, labeling on the second chromosome that should be marked by ChrS07 and ChrS09 (empty red circles) are missing. These are present in the lower cell. In the lower cell the second chromosomes of pairs 10 and 20 are not labeled (empty red circles), but they appeared in the upper cell. Scale bars = 5 µm; (c) Enumeration of S. intermedia chromosomes and their labeling by S. polyrhiza chromosome-specific probes.
Rights and permissions
About this article
Cite this article
Hoang, P.T.N., Schubert, I. Reconstruction of chromosome rearrangements between the two most ancestral duckweed species Spirodela polyrhiza and S. intermedia . Chromosoma 126, 729–739 (2017). https://doi.org/10.1007/s00412-017-0636-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-017-0636-7