Abstract
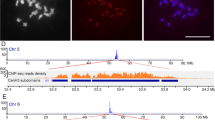
CENH3 is a centromere-specific histone H3 variant and has been used as a marker to identify active centromeres and DNA sequences associated with functional centromere/kinetochore complexes. In this study, up to four distinct CENH3 (BrCENH3) cDNAs were identified in individuals of each of three diploid species of Brassica. Comparison of the BrCENH3 cDNAs implied three related gene families: BrCENH3-A in Brassica rapa (AA), BrCENH3-B in B. nigra (BB), and BrCENH3-C in B. oleracea (CC). Each family encoded a histone fold domain and N-terminal histone tails that vary in length in all three families. The BrCENH3-B cDNAs have a deletion of two exons relative to BrCENH3-A and BrCENH3-C, consistent with the more ancient divergence of the BB genome. Chromatin immunoprecipitation and immunolabeling tests with anti-BrCENH3 antibodies indicated that both centromeric tandem repeats and the centromere-specific retrotransposons of Brassica are directly associated with BrCENH3 proteins. In three allotetraploid species, we find either co-transcription of the BrCENH3 genes of the ancestral diploid species or gene suppression of the BrCENH3 from one ancestor. Although B genome centromeres are occupied by BrCENH3-B in the ancestral species B. nigra, in allotetraploids both BrCENH3-A and BrCENH3-C proteins appear to assemble at these centromeres.








Similar content being viewed by others
References
Amor DJ, Choo KH (2002) Neocentromeres: role in human disease, evolution, and centromere study. Am J Hum Genet 71:695–714
Baker RE, Rogers K (2006) Phylogenetic analysis of fungal centromere H3 proteins. Genetics 174:1481–1492
Beilstein MA, Nagalingum NS, Clements MD, Manchester SR (2010) Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci USA 107:18724–18728
Black BE, Bassett EA (2008) The histone variant CENP-A and centromere specification. Curr Opin Cell Biol 20:91–100
Burnette WN (1981) ‘Western blotting’: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem 112:195–203
Bowers JE, Chapman BA, Rong JK, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438
Cheo TY, Lu LL, Yang G, Ihsan A, Shehbaz AL (2001) Flora of China 8. Science Press, Beijing
Cheung F, Trick M, Drou N, Lim YP, Park JY, Kwon SJ, Kim JA, Scott R, Pires JC, Paterson AH, Town C, Bancroft I (2009) Comparative analysis between homoeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. Plant Cell 21:1912–1928
Comeron JM (1999) K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15:763–764
Damerval C, Vienne DD, Zivy M, Thiellement H (1986) Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat seedling proteins. Electrophoresis 7:52–54
Dawe RK, Henikoff S (2006) Centromeres put epigenetics in the driver’s seat. Trends Biochem Sci 31:662–669
Fishman L, Saunders S (2008) Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322:1559–1562
Flagel L, Udall J, Nettleton D, Wendel J (2008) Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol 6:16
Furuyama T, Henikoff S (2009) Centromeric nucleosomes induce positive DNA supercoils. Cell 138:104–113
Gómez-Campo C, Prakash S (1999) Biology of brassica coenospecies. In: Gomez-Campo C (ed) In developments in plant genetics and breeding. Elsevier, Amsterdam, pp 33–52
Guyot R, Keller B (2004) Ancestral genome duplication in rice. Genome 47:610–614
Han YH, Wang GX, Liu Z, Liu JH, Yue W, Song RT, Zhang XY, Jin WW (2010) Divergence in centromere structure distinguishes related genomes in Coix lacryma-jobi and its wild relative. Chromosoma 119:89–98
Harrison GE, Heslop-Harrison JS (1995) Centromeric repetitive DNA sequences in the genus Brassica. Theor Appl Genet 90:157–165
Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098–1102
Heun P, Erhardt S, Blower M, Weiss S, Skora AD, Karpen GH (2006) Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell 10:303–315
Hirsch CD, Wu YF, Yan HH, Jiang JM (2009) Lineage-specific adaptive evolution of the centromeric protein CENH3 in diploid and allotetraploid Oryza species. Mol Biol Evol 26:2877–2885
Houben A, Schroeder-Reiter E, Nagaki K, Nasuda S, Wanner G, Murata M, Endo TR (2007) CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma 116:275–283
Hovav R, Udall JA, Chaudhary B, Rapp R, Flagel L, Wendell JF (2008) Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc Natl Acad Sci USA 105:6191–6195
Hudakova S, Michalek W, Presting GG, Hoopen RT, Santos KD, Jasencakova Z, Schubert I (2001) Sequence organization of barley centromeres. Nucleic Acids Res 29:5029–5035
Huson DH, Richter DC, Rausch D, Dezulian T, Franz M, Rupp R (2007) Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinform 22:460
Jasencakova Z, Soppe WJ, Meister A, Gernand D, Turner BM (2003) Histone modifications in Arabidopsis—high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J 33:471–480
Jiang JM, Nasuda S, Dong FG, Scherrer CW, Woo SS, Wing RA, Gill BS, Ward DC (1996) A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc Nat Acad Sci USA 93:14210–14213
Jin WW, Melo JR, Nagaki K, Talbert PB, Henikoff S, Dawe RK, Jiang JM (2004) Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell 16:571–581
Jin WW, Lamb JC, Zhang WL, Kolano B, Birchler JA, Jiang JM (2008) Histone modifications associated with both A and B chromosomes of maize. Chromosome Res 16:1203–1214
Ketel C, Helen S, Wang W, Mcclellan M, Bouchonville K, Selmecki A, Lahav T, Nejad MG, Berman J (2009) Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet 5:e1000400
Lagercrantz U, Lydiate DJ (1996) Comparative genome mapping in Brassica. Genetics 144:1903–1910
Lee HR, Zhang WL, Langdon T, Jin WW, Yan HH, Cheng ZK, Jing JM (2005) Chromatin immuno-precipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci USA 102:11793–11798
Lim KB, Jong HD, Yang TJ, Park JY, Kwon SJ, Kim JS, Lim MH, Kim JA, Jin M, Jin YM, Kim SH, Lim YP, Bang JA, Kim HI, Park BS (2005) Characterization of rDNAs and tandem repeats in the heterochromatin of Brassica rapa. Mol Cells 19:436–444
Lim KB, Yang TJ, Hwang YJ, Kim JS, Park JY, Kwon SJ, Kim JA, Beom SC, Lim MH, Jin M, Kim HI, Jong HD, Bancroft I, Lim YP, Park BS (2007) Characterization of the centromere and peri-centromere retrotransposons in Brassica rapa and their distribution in related Brassica species. Plant J 49:173–183
Liu Z, Yue W, Li DY, Wang R, Kong XY, Lu K, Wang GX, Dong YS, Jin WW, Zhang XY (2008) Structure and dynamics of retrotransposons at wheat centromeres and pericentromeres. Chromosoma 117:445–456
Lomiento M, Jiang ZS, Addabbo PD, Eichler EE, Rocchi M (2008) Evolutionary-new centromeres preferentially emerge within gene deserts. Genome Biol 9:R173
Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassiceae. Genome Res 15:516–525
Malik HS, Henikoff S (2001) Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157:1293–1298
Malik HS, Vermaak D, Henikoff S (2002) Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc Natl Acad Sci USA 99:1449–1454
Marmagne A, Brabant P, Thiellement H, Alix K (2010) Analysis of gene expression in resynthesized Brassica napus allotetraploids: transcriptional changes do not explain differential protein regulation. New Phytol 186:216–227
Marshall O, Chuel A, Wong L, Choo K (2008) Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Chromosoma 82:261–282
Nagaki K, Murata M (2005) Characterization of CENH3 and centromere-associated DNA sequences in sugarcane. Chromosome Res 13:195–203
Nagaki K, Cheng ZK, Yang SO, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang JM (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36:138–145
Nagaki K, Kashihara K, Murata M (2009) A centromeric DNA sequence colocalized with a centromere-specific histone H3 in tobacco. Chromosoma 118:249–257
Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR (2005) Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci USA 102:9842–9847
Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR (2007) CENP-B controls centromere formation depending on the chromatin context. Cell 131:1287–1300
Otto SP, Whitton J (2000) Polyploid incidence and evolution. Annu Rev Genet 34:401–437
Palmer DK, O’Day K, Trong HL, Charbonneau H, Margolis RL (1991) Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci USA 88:3734–3738
Parkin IA, Sharpe AG, Keith DJ, Lydiate DJ (1995) Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38:1122–1131
Parkin IA, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn T, Lydiate DJ (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171:765–781
Pignatta D, Comai L (2009) Parental squabbles and genome expression: lessons from the polyploids. J Biol 8:43
Rakow G (2004) Biotechnology in agriculture and forestry. In: Pua EC, Douglas CJ (eds) Species origin and economic importance of Brassica, in Brassica. Springer, Berlin, pp 3–11
Ravi M, Kwong PN, Menorca RM, Valencia JT, Ramahi JS, Stewart JL, Tran RK, Sundaresan V, Comai L, Chan SW (2010) The rapidly evolving centromere-specific histone has stringent functional requirements in Arabidopsis thaliana. Genetics 186:461–471
Schueler MG, Swanson W, Thomas PJ (2010) Adaptive evolution of foundation kinetochore proteins in primates. Mol Biol Evol 27:1585–1597
Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Sankoff D, dePamphilis CW, Wall PK, Soltis PS (2009) Polyploidy and angiosperm diversification. Am J Bot 96:336–348
Song K, Lu P, Osborn C (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci USA 92:7719–7723
Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11:1076–1083
Swigonova Z, Lai J, Ma J, Ramakrishna W, Llaca M, Bennetzen JL, Messing J (2004) On the tetraploid origin of the maize genome. Comp Funct Genom 5:281–284
Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S (2002) Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14:1053–1066
Talbert PB, Bryson TD, Henikoff S (2004) Adaptive evolution of centromere proteins in plants and animals. J Biol 3:18
Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics; Chapter 2: Unit 2.3
UN (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7:389–452
Vermaak D, Hayden HS, Henikoff S (2002) Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol 22:7553–7561
Wang GX, Zhang XY, Jin WW (2009) An overview of plant centromeres. J Genet Genomics 36:529–537
Warwick SI, Black LD, Aguinagalde I (1991) Molecular systematics of Brassica and allied genera (Subtribe Brassicinae, Brassiceae) chloroplast DNA variation in the genus Diplotaxis. Theor Appl Genet 83:839–850
Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P (2004) Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol Cell Biol 24:6620–6630
Wong NC, Wong LH, Quach JM, Canham P, Cragi JM, Song JZ, Clark SJ, Choo KHA (2006) Permissive transcriptional activity at the centromere through pockets of DNA hypomethylation. PLoS Genet 2:e17
Xu Y, Zhong L, Wu X, Fang X, Wang J (2009) Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta 229:471–483
Yan HH, Ito H, Nobuta K, Yang SO, Jin WW, Tian SL, Lu C, Venu RC, Wang GL, Green PJ, Wing RA, Buell CR, Meyers BC, Jiang JM (2006) Genomic and genetic characterization of rice Cen3 reveals extensive transcription and evolutionary implications of a complex centromere. Plant Cell 18:2123–2133
Yan HH, Talbert PB, Lee HR, Jett J, Henikoff S, Chen F, Jiang J (2008) Intergenic locations of rice centromeric chromatin. PLoS Biol 6:e286
Yang TJ, Kim JS, Kwon SJ, Lim KB, Choi BS, Kim JA, Jin M, Park JY, Lim MH, Kim HI, Lim YP, Kang JJ, Hong JH, Kim CB, Bhak J, Bancroft I, Park BS (2006) Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 18:1339–1347
Zhang WL, Lee HR, Koo DH, Jiang JM (2008) Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and Maize. Plant Cell 20:25–34
Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, Nagaki K, Birchler JA, Jiang JM, Dawe KR (2002) Centromeric retroelements and satellites interact with Maize kinetochore protein CENH3. Plant Cell 14:2825–2836
Acknowledgments
This research was supported by the Natural Science Foundation of China (31025018), by the Ministry of Science and Technology (2011CB944600), and by the Program for National Transgenic Research Program of China (2008ZX08009-001; 2009ZX08010-003B) to W.W. Jin.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by L. Comai
Guixiang Wang and Qunyan He contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
RACE sequences from B. rapa cultivar Beijingxin2 (AA). 5-RACE generated 5 clones representing two sequences, A2 and A3. 3-RACE generated 3 clones with a single sequence. The positions of 5′ and 3′ RACE primers are underlined. The start and stop codons of the open reading frame are in bold font (DOC 26 kb)
Fig. S2
Alignment of Brassica cDNAs. Brassica cDNAs comprise three large groups (BrCENH3-A, BrCENH3-B, and BrCENH3-C) differentiated by common nucleotide substitutions and length variations. Nucleotides that differ from the consensus are marked: BrCENH3-A—pink, BrCENH3-B—blue, BrCENH3-C—yellow. The primers used to amplify the cDNAs are shown in gray. The regions of the cDNA clones corresponding to the primers may not reflect exactly the sequences of the underlying genes except for A2 and A3, which represent complete coding sequences of BrCENH3-1 as determined by RACE. Inferred intron positions are marked with |. The predicted coding sequences from four genomic sequences from B. juncea (AB_AB299179 and AB_AB299179) and B. oleracea (C_AB299276 and C_AB299276) are included for reference (DOC 64 kb)
Fig. S3
Genomic sequences from Brassica rapa. Genomic sequences from the BrCENH3-A coding regions of B. rapa accessions Beijingxin2, Jingchunbai98-7, or from both accessions. The genomic sequence of the BrCENH3-A gene of B. juncea (AB299179) is shown for comparison. Overlines indicate the location of PCR primers. Exons are in blue type, and introns are in black type. Alternative donor and acceptor splice sites bordering the third intron are in bold italics (DOC 42.5 kb)
Fig. S4
The Prokaryotic Expression of pGEX-4T-2+BrCENH3. M, protein marker; 1, uninduced bacteria transfected with pGEX-4T-2; 2, bacteria (transfected with pGEX-4T-2) after induction with IPTG; 3 and 5, uninduced bacteria Rosetta transfected with pGEX-4T+BrCENH3; 4 and 6, Bacteria Rosetta (transfected with pGEX-4T+BrCENH3) after induction with IPTG (DOC 206 kb)
Supplementary Table S1
Primers for amplying CENH3 in this study (DOC 44 kb)
Rights and permissions
About this article
Cite this article
Wang, G., He, Q., Liu, F. et al. Characterization of CENH3 proteins and centromere-associated DNA sequences in diploid and allotetraploid Brassica species. Chromosoma 120, 353–365 (2011). https://doi.org/10.1007/s00412-011-0315-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-011-0315-z