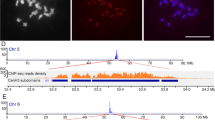
Abstract
Knowledge about the composition and structure of centromeres is critical for understanding how centromeres perform their functional roles. Here, we report the sequences of one centromere-associated bacterial artificial chromosome clone from a Coix lacryma-jobi library. Two Ty3/gypsy-class retrotransposons, centromeric retrotransposon of C. lacryma-jobi (CRC) and peri-centromeric retrotransposon of C. lacryma-jobi, and a (peri)centromere-specific tandem repeat with a unit length of 153 bp were identified. The CRC is highly homologous to centromere-specific retrotransposons reported in grass species. An 80-bp DNA region in the 153-bp satellite repeat was found to be conserved to centromeric satellite repeats from maize, rice, and pearl millet. Fluorescence in situ hybridization showed that the three repetitive sequences were located in (peri-)centromeric regions of both C. lacryma-jobi and Coix aquatica. However, the 153-bp satellite repeat was only detected on 20 out of the 30 chromosomes in C. aquatica. Immunostaining with an antibody against rice CENH3 indicates that the 153-bp satellite repeat and CRC might be both the major components for functional centromeres, but not all the 153-bp satellite repeats or CRC sequences are associated with CENH3. The evolution of centromeric repeats of C. lacryma-jobi during the polyploidization was discussed.





Similar content being viewed by others
References
Amor DJ, Choo KH (2002) Neocentromeres: role in human disease, evolution, and centromere study. Am J Hum Genet 71:695–714
Ananiev EV, Phillips RL, Rines HW (1998) Chromosome specific molecular organization of maize (Zea mays L.) centromeric regions. Proc Natl Acad Sci USA 95:13073–13078
Arora RK (1977) ‘Job’s tears’(Coix lacryma-jobi), a minor food and fodder crop of North-eastern India. Economic Botany 31:358–366
Barve SS, Spare AB (1986) Mono-trisomic in Coix gigantea. Curr Sci 55:660–661
Cheng Z, Dong F, Langdon T, Ouyang S, Buell CR, Gu M, Blattner FR, Jiang J (2002) Functional rice centromeres are marked by a satellite repeat and a centromerespecific retrotransposon. Plant Cell 14:1691–1704
Christopher J, Benny J (1990) Cytology of a new Hexaploid cyto-type of Coix lacryma-jobi Linn. Cytologia 55:57–60
Clayton WD (1981) Notes on tribe Andropogoneae (Graminea). Rew Bulletin 35:813–818
Dalal Y, Wang H, Lindsay S, Henikoff S (2007) Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol 5:e218
Dong F, Miller JT, Jackson SA, Wang GL, Ronald PC, Jiang J (1998) Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc Natl Acad Sci USA 95:8135–8140
Fitzgerald DJ, Dryden GL, Bronson EC, Williams JS, Anderson JN (1994) Conserved patterns of bending in satellite and nucleosome positioning DNA. J Biol Chem 269:21303–21314
Hall SE, Kettler G, Preuss D (2003) Centromere satellites from Arabidopsis populations: maintenance of conserved and variable domains. Genome Res 13:195–205
Han YH, Li DY, Li YC, Xue YG, Hu ZL, Song YC (2004) Cytogenetic identification and analysis of a new hexaploid Coix aquatica Roxb. cyto-type. Acta Botanica Sinica 46:724–729
Henikoff S (2002) Near the edge of a chromosome’s ‘black hole’. Trends Genet 18:165–167
Henikoff S, Dalal Y (2005) Centromeric chromatin: what makes it unique? Curr Opin Genet Dev 15:177–184
Houben A, Brandes A, Pich U, Manteuffel R, Schubert I (1996) Molecular-cytogenetic characterization of a higher plant centromere/kinetochore complex. Theor Appl Genet 93:477–484
Houben A, Schroeder-Reiter E, Nagaki K, Nasuda S, Wanner G, Murata M, Endo TR (2007) CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma 116:275–283
Hudakova S, Michalek W, Presting GG, ten Hoopen R, dos Santos K, Jasencakova Z, Schubert I (2001) Sequence organization of barley centromeres. Nucleic Acids Res 29:5029–5035
Jackson SA, Wang ML, Goodman HM, Jiang J (1998) Application of Fiber-FISH in genome analysis of Arabidopsis thaliana. Genome 41:566–572
Jackson SA, Dong F, Jiang J (1999) Digital mapping of bacterial artificial chromosomes by fluorescence in situ hybridization. Plant J 17:581–587
Jiang J, Gill BS, Wang GL, Ronald PC, Ward DC (1995) Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci 92:4487–4491
Jiang J, Birchler JA, Parrott WA, Dawe RK (2003) A molecular view of plant centromeres. Trends Plant Sci 8:570–575
Jin W, Melo JR, Nagaki K, Talbert PB, Henikoff S, Dawe RK, Jiang J (2004) Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell 16:571–581
Jin W, Lamb JC, Vega JM, Dawe RK, Birchler JA, Jiang J (2005) Molecular and functional dissection of the maize B chromosome centromere. Plant Cell 17:1412–1423
Kellogg EA (2001) Evolutionary history of the grasses. Plant Physiol 125:1198–1205
Kong XY, Gu YQ, You FM, Dubcovsky J, Anderson OD (2004) Dynamics of the evolution orthologous and paralogous portions of a complex locus region in two genomes of allopolyploid wheat. Plant Mol Biol 54:55–69
Kulikova O, Gualtieri G, Geurts R, Kim DJ, Cook D, Huguet T, de Jong JH, Fransz PF, Bisseling T (2001) Integration of the FISH pachytene and genetic maps of Medicago truncatula. Plant J 27:49–58
Kulikova O, Geurts R, Lamine M, Kim DJ, Cook DR, Leunissen J, de Jong H, Roe BA, Bisseling T (2004) Satellite repeats in the functional centromere and pericentromeric heterochromatin of Medicago truncatula. Chromosoma 113:276–283
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Kumekawa N, Ohtsubo H, Horiuchi T, Ohtsubo E (1999) Identification and characterization of novel retrotransposons of the gypsy type in rice. Mol Gen Genet 260:593–602
Kumekawa N, Hosouchi T, Tsuruoka H, Kotani H (2000) The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 5. DNA Res 7:315–321
Lamb JC, Theuri J, Birchler JA (2004) What’s in a centromere? Genome Bio 5:239
Langdon T, Seago C, Mende M, Leggett M, Thomas H, Forster JW, Jones RN, Jenkins G (2000) Retrotransposon evolution in diverse plant genomes. Genetics 156:313–325
Lee HR, Zhang W, Langdon T, Jin W, Yan H, Cheng Z, Jiang J (2005) Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci USA 102:11793–11798
Li YX, Kirby ML (2003) Coordinated and conserved expression of alphoid repeat and alphoid repeat-tagged coding sequences. Dev Dynamics 228:72–81
Lim KB, de Jong H, Yang TJ, Park JY, Kwon SJ, Kim JS, Lim MH, Kim JA, Jin M, Jin YM, Kim SH, Lim YP, Bang JW, Kim HI, Park BS (2005) Characterization of rDNAs and tandem repeats in heterochromatin of Brassica rapa. Mol Cells 19:436–444
Lim KB, de Jong H, Yang TJ, Park JY, Kwon SJ, Kim JS, Lim MH, Kim JA, Jin M, Jin YM, Kim SH, Lim YP, Bang JW, Kim HI, Park BS (2007) Characterization of the centromere and peri-centromere retrotransposons in Brassica rapa and their distribution in related Brassica species. Plant J 49:173–183
Lin JY, Jacobus BH, SanMiguel P, Walling JG, Yuan Y, Shoemaker RC, Young ND, Jackson SA (2005) Pericentromeric regions of soybean (Glycine max L. Merr.) chromosomes consist of retroelements and tandemly repeated DNA and are structurally and evolutionarily labile. Genetics 170:1221–1230
Liu Z, Yue W, Li D, Wang R, Kong XY, Kun Lu, Wang GX, Dong YS, Jin WW, Zhang XY (2008) Structure and dynamics of retrotransposons at wheat centromeres and pericentromeres. Chromosoma 117:445–456
Lu P, Zuo ZM (1996) Detection and identification of Coix aquatica species in Guangxi. Guangxi Agric Sci 1:18–20
Ma J, Wing RA, Bennetzen JL, Jackson SA (2007) Plant centromere organization: a dynamic structure with conserved functions. Trends Genet 23:134–139
Malik HS, Bayes JJ (2006) Genetic conflicts during meiosis and the evolutionary origins of centromere complexity. Biochem Soc Trans 34:569–573
Malik HS, Henikoff S (2002) Conflict begets complexity: the evolution of centromeres. Curr Opin Genet Dev 12:711–718
Martínez-Balbás A, Rodríguez-Campos A, García-Ramírez M, Sainz J, Carrera P, Aymamí J, Azorín F (1990) Satellite DNAs contain sequences that induce curvature. Biochemistry 29:2342–2348
Miller JT, Dong F, Jackson SA, Song J, Jiang J (1998) Retrotransposon-related DNA sequences in the centromeres of grass chromosomes. Genetics 150:1615–1623
Mravinac B, Plohl M, Ugarković Ð (2004) Conserved patterns in the evolution of Tribolium satellite DNAs. Gene 332:169–177
Mravinac B, Plohl M, Ugarković Ð (2005) Preservation and high sequence conservation of satellite DNAs suggest functional constraints. J Mol Evol 61:542–550
Nagaki K, Murata M (2005) Characterization of CENH3 and centromere-associated DNA sequences in sugarcane. Chromosome Res 13:195–203
Nagaki K, Song J, Stupar RM, Parokonny AS, Yuan Q, Ouyang S, Liu J, Hsiao J, Jones KM, Dawe RK, Buell CR, Jiang J (2003) Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics 163:759–770
Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36:138–145
Presting GG, Malysheva L, Fuchs J, Schubert I (1998) A Ty3/gypsy retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J 16:721–728
Round EK, Flowers SK, Richards EJ (1997) Arabidopsis thaliana centromere regions: genetic map positions and repetitive DNA structure. Genome Res 7:1045–1053
Schubert I (1998) Late-replicating satellites: something for all centromeres? Trends Genet 14:385–387
Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF (2001) Genomic and genetic definition of a functional human centromere. Science 294:109–115
Thompson H, Schmidt R, Brandes A, Heslop-Harrison JS, Dean C (1996) A novel repetitive sequence associated with the centromeric regions of Arabidopsis thaliana chromosomes. Mol Gen Genet 253:247–252
Ugarkovic DL, Plohl M (2002) Variation in satellite DNA profiles—causes and effects. EMBO J 21:5955–5959
Ugarkovic DL, Plohl M, Lucijanic-Justic V, Borstnik B (1992) Detection of satellite DNA in Palorus ratzeburgii: analysis of curvature profiles and comparison with Tenebrio molitor satellite DNA. Biochimie 74:1075–1082
Ugarkovic DL, Podnar M, Plohl M (1996) Satellite DNA of the red flour beetle Tribolium castaneum-comparative study of satellites from the genus Tribolium. Mol Biol Evol 13:1059–1066
Wicker T, Stein N, Albar L, Feuillet C, Schlagenhauf E, Keller B (2001) Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J 26:307–316
Woo JH, Li D, Wilsbach K, Orita H, Coulter J, Tully E, Kwon TK, Xu S, Gabrielson E (2007) Coix seed extract, a commonly used treatment for cancer in China, inhibits NFkappaB and protein kinase C signaling. Cancer Biol Ther 6:2005–2011
Yang TJ, Lee S, Chang SB, Yu Y, de Jong H, Wing RA (2005) In-depth sequence analysis of the tomato chromosome 12 centromeric region: identification of a large CAA block and characterization of pericentromere retrotranposons. Chromosoma 114:103–117
Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, Nagaki K, Birchler JA, Jiang J, Dawe RK (2002) Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14:2825–2836
Acknowledgments
The authors are grateful to Dr. J.S. Li (China Agricultural University) for supplying the seeds of Coix. This research was supported by grant (30771208) from the National Science Foundation, State Key Basic Research and Development Plan of China (973; 2009CB118400), and National Transgenic Research Program of China (2008ZX08009-001).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by: I. Schubert
Yonghua Han and Guixiang Wang made an equal contribution to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Analysis of chromosome distribution and insert sizes of representative BAC clones 9-6-12 and 9-18-39. (a) FISH analysis of BAC 9-6-12. The signals dispersed along all chromosomes with an increased concentration around centromeric regions. (b) FISH analysis of BAC 9-18-39. The signals were exclusively detected in centromeres. (c) Insert sizes and fingerprint of BAC 9-6-12(12) and 9-18-39(39). After restriction of NotI and removing the 7.5 kb-length vector, the insert sizes were ∼130 and 110 kb, respectively. BAC 12 and 39 were not in the same contig as revealed in fingerprint by restriction with HindIII (GIF 72 kb)
Supplementary Fig. 2
Sequence comparison between 20 variant monomers of the 153-bp satellite repeat. Sequence conservation is indicated by background shading, where black represents 100% identity, dark gray represents at least 80%, and light gray represents at least 60%. Asterisks represent nucleotide position 10, 30, 50, and 70, respectively (GIF 1261 kb)
Supplementary Fig. 3
Highly conserved sequence motifs identified in the LTRs of the centromere-specific retrotransposons in the grass species were found in the LTRs of CRC (GIF 5275 kb)
Rights and permissions
About this article
Cite this article
Han, Y., Wang, G., Liu, Z. et al. Divergence in centromere structure distinguishes related genomes in Coix lacryma-jobi and its wild relative. Chromosoma 119, 89–98 (2010). https://doi.org/10.1007/s00412-009-0239-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-009-0239-z