Abstract
In mammals, chromosomes occupy defined positions in sperm, whereas previous work in chicken showed random chromosome distribution. Monotremes (platypus and echidnas) are the most basal group of living mammals. They have elongated sperm like chicken and a complex sex chromosome system with homology to chicken sex chromosomes. We used platypus and chicken genomic clones to investigate genome organization in sperm. In chicken sperm, about half of the chromosomes investigated are organized non-randomly, whereas in platypus chromosome organization in sperm is almost entirely non-random. The use of genomic clones allowed us to determine chromosome orientation and chromatin compaction in sperm. We found that in both species chromosomes maintain orientation of chromosomes in sperm independent of random or non-random positioning along the sperm nucleus. The distance of loci correlated with the total length of sperm nuclei, suggesting that chromatin extension depends on sperm elongation. In platypus, most sex chromosomes cluster in the posterior region of the sperm nucleus, presumably the result of postmeiotic association of sex chromosomes. Chicken and platypus autosomes sharing homology with the human X chromosome located centrally in both species suggesting that this is the ancestral position. This suggests that in some therian mammals a more anterior position of the X chromosome has evolved independently.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
A rapidly increasing body of evidence shows that higher-order organization in the nucleus is essential for a genome to function properly (Cremer et al. 2001; Fraser and Bickmore 2007). A general theme of genome organization in the interphase nucleus is that gene-rich chromosomes are organized preferentially towards the interior of the nucleus whereas gene-poor chromosomes are located more towards the exterior (Cremer et al. 2001).
One of the most obvious differences in genome organization between mammals and reptiles is the presence of a distinct set of small very gene-rich microchromosomes (MICs) and large gene-poor macrochromosomes (McQueen et al. 1996, 1998). In interphase nuclei, MICs are found in the nuclear interior, while macrochromosomes (MACs) reside towards the nuclear periphery (Habermann et al. 2001). Monotremes are the only mammalian species that may share microchromosomes with reptiles (Matthey 1949; White 1973). However, this suggestion was based on chromosomes size alone and has been challenged by Van Brink as both platypus and echidna show a continuum of chromosome size, while in reptiles there is a bimodal distribution of chromosomes that clearly separate as MICs and MACs (Van Brink 1959). Recently, we described the localization of MHC genes on the smallest platypus chromosomes, which show homology to chicken MICs 16 and 17 (Dohm et al. 2007).
Sex chromosome systems are different in birds and mammals. In chicken, the heterogametic sex is the female (ZW), whereas in mammals it is the male (XY). Male chickens have two Z chromosomes and female mammals two X chromosomes. Comparative mapping revealed that the chicken Z and mammalian X do not share homology and most of the genes from the eutherian X are found on chicken chromosomes 1 and 4 (Kohn et al. 2007; Nanda et al. 1999). The platypus (Ornithorhynchus anatinus) sex chromosome system is remarkably different to both therian mammals and birds in that there are ten sex chromosomes that form a meiotic chain in prophase I (Grutzner et al. 2004; Rens et al. 2004). The sex chromosomes of the chain undergo alternate segregation to form male-determining sperm with five Y chromosomes and female-determining sperm with five X chromosomes (Grutzner et al. 2004). Surprisingly, the platypus sex chromosome system shows homology with the chicken Z (Grutzner et al. 2004). The recent sequencing of the entire platypus genome revealed that there is no homology between the mammalian X, and most of the genes from the human X that have been mapped in platypus reside on chromosome 6 (Veyrunes et al. 2008; Warren et al. 2008; Waters et al. 2005). Another unusual feature of monotreme sex chromosomes is that they contain MHC genes. The major histocompatibility complex (MHC) is of central importance for adaptive and innate immunity in vertebrates. Sequencing MHCs from different mammalian species have led to the identification of approximately 220 genes located within an interval of 3.5–4 Mbp, which have important roles for example in immunity and reproduction. In human, the 3.6-Mb MHC complex is located on chromosome 6 (Horton et al. 2004). We have recently mapped MHC genes on platypus Y3X3 and Y4X5 (Dohm et al. 2007). In chicken, the MHC is much smaller. The MHC class I and class II genes are found on GGA 16, while framework genes that localize to the human class III interval map to GGA 17 (Fillon et al. 1998).
In mammals, sperm have evolved various shapes and contain elaborate acrosome and tail structures for efficient motility and fertilization. It is generally thought that vermiform sperm found in many reptile species and monotremes represent less sophisticated, ancestral sperm morphology (Bedford 1991, 2004).
In terms of genome organization, non-random distribution of almost all chromosomes has been demonstrated in several mammalian species (Foster et al. 2005; Greaves et al. 2003, 2001; Haaf and Ward 1995; Hazzouri et al. 2000; Meyer-Ficca et al. 1998; Ward et al. 1996; Zalenskaya and Zalensky 2004). Human chromosome 17 for example, which is one of the gene-richest chromosomes, located consistently in the middle of the sperm nucleus. In contrast, the gene-poor human chromosome 13 was found randomly distributed within the sperm nucleus (Hazzouri et al. 2000; Zalenskaya and Zalensky 2004).
Monotremes diverged more than 160 million years ago from the mammalian lineage and are therefore the oldest branch on the mammalian tree (Bininda-Emonds et al. 2007). The elongated sperm in monotremes has a similar structure to that of birds and reptiles and therefore they retain the ancestral structure (Bedford 1991). A thorough comparison of the different stages of platypus spermiogenesis has indeed identified similarities but also differences between platypus and reptilian sperm development and morphology (Lin and Jones 2000). For platypus, earlier work showed tandem arrangement of telomeric sequences in sperm and localized ribosomal DNA (rDNA) and genes from platypus chromosome 6 to specific regions in sperm. The mapping of a whole-chromosome paint of platypus X1 to the anterior of platypus sperm provided additional evidence to show a non-random organization of chromosomes in platypus sperm (Greaves et al. 2003; Watson et al. 1996).
In chicken, previous research on the arrangement of centromeric heterochromatin found some evidence for ordered arrangement in sperm (Dressler and Schmid 1976). In contrast, more recent work reported that chicken MACs GGA 2, GGA 6, GGA 8, GGA 9, and GGA Z and a single MIC show random organization in chicken sperm (Greaves et al. 2003; Solovei et al. 1998). In addition, Greaves et al. (2003) utilized the fact that telomere repeats show a stronger signal on MICs (Nanda and Schmid 1994) to demonstrate that some MICs apparently cluster in the central region of sperm.
The evolution of heteromorphic sex chromosomes is accompanied by the establishment of regional dosage compensation in birds (Melamed and Arnold 2007) or chromosome-wide dosage compensation mechanisms in the homogametic sex in birds and mammals and meiotic sex chromosome silencing in the heterogametic sex in mammals (Heard and Disteche 2006). How both epigenetic mechanisms relate to genome organization in sperm is unknown. The random positioning of the chicken Z seems to rule out any importance of position in sperm for dosage compensation. In mammals, however, a high conservation of the mammalian X chromosome in a position that may make first contact with the oocyte has led to the idea that the position of the X chromosome could play a role in the fertilization process or may be important for imprinted X inactivation (Greaves et al. 2003). A more internal location of the X in pigs is not compatible with this idea (Foster et al. 2005). Apart from the mammalian X chromosome little is known about the evolutionary conservation of chromosome positioning in sperm. This is partly due to the fact that sperm shapes differ significantly between species, making comparisons difficult. Monotremes are one of only a few mammals that share elongated sperm with chicken and other reptiles (Fig. 1), which allows a more precise comparison of the genome organization between mammals and birds.
The main function of sperm is to transport the paternal DNA and to successfully fertilize an egg. In contrast to the interphase nucleus, very few genes are actively transcribed in sperm, and during spermatogenesis, the chromatin is reorganized extensively by the replacement of most of the histone proteins by transitional proteins and later protamines to allow for a more efficient compaction in the sperm nucleus (Kimmins and Sassone-Corsi 2005). In humans, chromosome territories were four times more condensed in sperm than in metaphase chromosomes (Mudrak et al. 2005). On the other hand, extended chromatin and chromosome territories have been observed in mammalian species like pig and rat (Foster et al. 2005; Meyer-Ficca et al. 1998) and also in chicken sperm (Solovei et al. 1998). Chromatin compaction is achieved by the positively charged arginine-rich protamines, which replace most of the histone proteins in sperm. Protamines are evolutionarily conserved in vertebrates including birds and monotremes. However, cysteine residues present in eutherian sperm are absent in pangolins (eutherian), marsupials, monotremes, birds, and fish, which make their sperm chromatin much more accessible as it is not stabilized with disulfide bridges (Leung and Cummins 1988; Oliva and Dixon 1989; Retief et al. 1993).
The comparison of genome organization in the very similar platypus and chicken sperm is ideally suited to discern if the differences that have been observed in the arrangement of chromosomes in chicken and mammalian sperm are due to differences in the genome (i.e., chromosome size, gene density, sex chromosome system) or differences in sperm morphology.
We have utilized the availability of chromosome-specific bacterial artificial chromosomes (BAC) clones from the chicken and platypus genome projects (Wallis et al. 2004; Warren et al. 2008). This enabled us to determine the position of several small and large autosomes and sex chromosomes in chicken and platypus sperm and to compare our results with existing data in sperm of eutherian mammals. In addition, we have quantified chromatin compaction using chicken BAC clones with known physical distance. Our findings reveal new aspects about genome organization in elongated chicken and platypus sperm and provide new insights into the evolution of chromosome organization in the sperm nucleus in birds and mammals.
Materials and methods
Preparation of chromosomes and meiotic cells
Mitotic metaphase chromosomes and interphase preparations were generated from the established platypus fibroblast cell lines (Grutzner et al. 2004). New cultures were set up from toe web from animals captured at the upper Barnard river (New South Wales, Australia) during breeding season (AEC permit no. S-49-2006 to F.G.). Meiotic cells and sperm were obtained from extracted testis. The material was either directly fixed in methanol/acetic acid (3:1) or incubated in 0.075 M KCl M at 37°C as hypotonic treatment and then fixed. Slides were prepared according to standard procedures. Chicken sperm was prepared from an adult male rooster the same way with all material subject to 0.075 M KCl hypotonic treatment.
BAC clones
Platypus BAC clones were obtained from the CUGI BAC/EST resource center (Clemson, SC, USA) from the platypus BAC library Oa_Ab or from the Children’s Hospital Oakland Research Institute (CHORI, CA, USA) from the platypus BAC library CH236 and chicken BAC library CH261. All BAC clones were mapped to metaphase chromosomes to confirm their location. For BAC clones from microchromosomes, we relied on the chicken genome sequence assignments from individual BAC clones as they cannot be distinguished cytogenetically (Table 1 contains a list of all BAC clones).
Fluorescence in situ hybridization of BAC clones
DNA (1 μg) from the positive BAC clones were directly labeled with spectrum orange or spectrum green (Vysis) using random primers and Klenow polymerase and hybridized by fluorescence in situ hybridization to platypus and chicken metaphase, interphase, spermatids, and sperm chromosomes under standard conditions. Briefly, the slides were treated with 100 μg/ml RNase A/2× SSC 37°C for 30 min and with 0.01% pepsin in 10 mM HCl at 37°C for 10 min. After re-fixing for 10 min in 1× PBS, 50 mM MgCl2, 1% formaldehyde, the preparations were dehydrated in an ethanol series. Slides were denatured for 2.5 min at 75°C in 70% formamide, 2× SSC, pH 7.0 and again dehydrated. For hybridization of one half slide, 400–500 ng of probe DNA was coprecipitated with 10–20 μg of boiled genomic platypus or echidna DNA (as competitor), and 50 μg salmon sperm DNA (as carrier), and redissolved in 50% formamide, 10% dextran sulfate, 2× SSC. The hybridization mixture was denatured for 10 min at 80°C. Preannealing of repetitive DNA sequences was carried out for 30 min at 37°C. The slides were hybridized overnight in a moist chamber at 37°C. The slides were then washed three times for 5 min in 50% formamide, 2× SSC at 42°C and once for 5 min in 0.1× SSC. Chromosomes and cell nuclei were counterstained with 1 μg/ml DAPI in 2× SSC for 1 min and mounted in 90% glycerol, 0.1 M Tris–HCl, pH 8.0 and 2.3% DABCO. Images were taken with a Zeiss AxioImagerZ.1 epifluorescence microscope equipped with a CCD camera and Zeiss Axiovision software.
Data analysis
Fluorescence in situ hybridization (FISH) signals were counted in 90 or more sperm where both ends could be clearly identified and the signal could be assigned to an anterior medial or posterior position. When several different clones were used, the results were added.
Measurements were taken with Olympus AnalySIS software. The distance of signals was derived from three measurements (see Fig. 2). In the AnalySIS software, a macro was prepared in which information about the magnification was entered and a scale bar in the picture was used to calibrate the measurements. The measurements were taken by manually drawing a line from each of the signals to the anterior end of the sperm, or along the whole sperm length (see Fig. 2). Excel was used to produce charts and perform ANOVA tests correlating signal distance and sperm length as well as for the chi-square analysis of the signal distribution with 2 degrees of freedom.
Identification of the clone position in sperm. Example of FISH signals in platypus sperm (a). Anterior, medial, and posterior were defined by dividing the total length of the sperm nucleus into three equal parts and assigning the signals by eye (b). Sperm length and signal distance were measured using Olympus AnalySIS software. To minimize variability, measurements were taken along the center of the sperm (c–e). The distance from the anterior tip of the sperm to the beginning of the first signal was measured (c), and the length of the line was recorded as ‘a’ μm. The line was continued through to the start of the second signal (d), and the total length of the line was recorded as ‘b’ μm. The line was continued from the beginning of the second signal to the posterior end of the sperm (e), and the length of the line was recorded as ‘c’ μm. The distance d between the signals was calculated as d = b − a
Results
Localization of chicken and platypus specific BAC clones in elongated sperm
In all experiments, we have hybridized chromosome-specific BAC clones to metaphase chromosomes and spermatids simultaneously. The position of signals was determined in more than 90 elongated sperm. The sperm nucleus was divided into three equal parts: anterior (pointed end), medial, and posterior, and each signal was assigned by eye to one of the three regions (Fig. 2). Table 1 lists the individual clones, their chromosome position, and the distribution in sperm. If more than one BAC was used from one chromosome, the data were merged (Tables 2 and 3). We investigated three aspects of genome organization in elongated chicken and platypus sperm, i.e., positioning and orientation of chromosomes and compaction of chromatin in sperm.
Higher-order genome organization is present in chicken sperm
In chicken (2n = 78, ZW female, ZZ male sex chromosome system), we have mapped 24 BAC clones from 12 different chicken (Gallus gallus) macro- and microchromosomes including the Z chromosome in elongated sperm (see Fig. 3 and Tables 1 and 2). GGA 1 and in GGA 4 are macrochromosomes and share extensive homology with the mammalian X chromosome (Kohn et al. 2007). We found a random distribution for two BAC clones from the proximal long arm of GGA 1. We observed a higher frequency of signals in the medial region of sperm nuclei (44.5%), but the overall distribution of signals along the sperm does not significantly deviate from a random distribution (Table 2). Two BAC clones from the short arm of the acrocentric chicken chromosome 4 showed a higher frequency of signals in the middle or towards the posterior of sperm and only 19% of signals were found in the anterior of the sperm (Fig. 3, Table 2). These results reveal a significant non-random distribution of GGA 4. For GGA 9, we saw a very even signal distribution and a clearly random organization. A previous study reported that the heterochromatic part of the chicken Z chromosome is randomly distributed in sperm (Solovei et al. 1998). Our results from mapping five BAC clones from different parts of the Z chromosome also show a random distribution. However, we noted very different distributions for the individual clones, where specific BACs reside with frequencies of up to 42% in all three parts of the sperm (see Table 1). For example clone CH261-66E20 (Zq) is found more than twice as frequent in the anterior than in the posterior part. The opposite is true for clones CH261-78C3 and CH261-89F20 (Zp) (Table 1). In general, we observed that the long arm of GGA Z seems to be located towards the anterior and the short arm of GGA Z towards the posterior of sperm. It is noteworthy that for GGA 1, GGA 4, and GGA Z individual BAC clones from these chromosomes differed significantly in their distribution along the sperm nucleus (see Table 1).
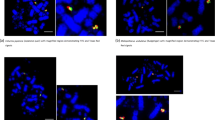
Mapping of chicken BACs in chicken cells. Using two-color FISH, we localized chromosomes in mitotic metaphase (a, d, g, h), interphase (b, e, h, k), and sperm (c, f, i, l). Examples are shown for BACs CH261-15M15 (114 Mb, green) and CH261-114L9 (134 Mb, red) from GGA 1, (a, b, c), BACs CH261-98D21 (10 Mb, red) and CH261-173H15 (4 Mb, green) from GGA 4 (d, e, f), BACs CH261-89F20 (23 Mb, red) and CH261-66E20 (58 Mb, green) GGA Z (g, h, i) and BACs CH261-128H20 (1.2 Mb, green) and CH261-18A6 (2.4 Mb, red) GGA 27 (j, k, l). Scale bar is 10 μm
Microchromosomes are the most euchromatic part of the chicken genome and contain over 50% of all chicken genes (Hillier et al. 2004). We have mapped BAC clones from eight different MICs onto sperm to look for their distribution. Interestingly, four MICs, 16, 17, 27, and 28, showed a clear non-random distribution with all chromosomes residing in over 43% of cases in the middle of sperm. The other MICs, 20, 23, 24, and 26, were randomly distributed although the highest signal frequencies were mostly seen in the middle of sperm (Table 2). To see if overall MICs tend to cluster in the middle as preliminary data suggested using telomere repeats (Greaves et al. 2003), we added all frequencies from MICs (29% anterior, 42% medial, 29% posterior). This shows a slightly (statistically not significant) higher value in the middle (42%). We conclude that there is no significant overrepresentation of MICs in the middle of sperm.
Non-random organization of autosomes and clustering of multiple sex chromosomes is present in platypus sperm
Both platypus and chicken share elongated sperm. In addition, platypus chromosomes show vastly different chromosome size as in birds and most other reptiles and a unique ten sex chromosome system.
In platypus (2n = 52, 21 autosomes, ten sex chromosomes), we have mapped 13 BAC clones from 12 different chromosomes in elongated sperm (Table 3). The largest platypus chromosomes, OAN 1, 2, and 3, showed a non-random position with chromosomes 1 and 3 in the anterior and chromosome 3 in medial regions of sperm. We were most interested to investigate the organization of the multiple sex chromosomes in sperm. We asked the following questions:
-
Do X and Y chromosomes show any association or clustering in sperm?
-
Do platypus sex chromosomes that share homology to GGA Z show an organization in sperm that is similar to chicken (random) or to other mammals (mostly anterior)?
-
Does the autosome OAN 6, which shares extensive homology with the human X reside in an anterior position as found for the X in several mammals or in the middle as we found for the homologous GGA 4?
To investigate the organization of the multiple X and Y chromosomes, we utilized a set of BAC clones from XY-shared regions as well as clones X- and Y-specific regions to distinguish between X- and Y-bearing sperm (see Fig. 4a).
Mapping of platypus BAC clones in platypus cells. Mitotic metaphase chromosomes (a, e, g), interphase cells (b, h), and sperm (c, d, f, i). In order to differentiate between X-bearing and Y-bearing sperm, combinations of BAC clones from the XY-shared and the X- and Y-specific regions were utilized in a two-color FISH experiment. For example, for X1Y1 BAC CH261-15M15 from X1pY1 (red) was hybridized together with BAC CH236-378F21 from X1q (green; a–d). Sperm with only the red signal are Y1 bearing (c) and sperm with red and green signal X1 bearing (d). Hybridization of a Y5-specific BAC Oa_Bb 425O3 in green (e). Sperm from different pictures have been assembled in this panel to illustrate the different signal shape. Note the elongated signal in some sperm (f). Localization of BAC CH236-577N02 (red) from the proximal long arm of OAN 6 and BAC CH236-97N06 (green) from the distal long arm (g). Two BAC clones in close proximity from OAN 6, Oa_Bb300C4 in red and Oa_Bb 425O3 in green, are shown in (h, insert). Despite the proximity of the clones in metaphase chromosomes and in interphase (h), they could be far apart in sperm (i). Scale bar is 10 μm
We used two BAC clones from X1p (XY shared) and X1q (X specific). Overall, we found that X1 resides in the medial region of platypus sperm. However, we observed that the BAC clone from X1p localized more frequently in the anterior region (39%) compared to the BAC clone from X1q (18%), which resides very consistently in the medial part of the sperm (52%). The combination of BAC clones from the XY-shared region (on X1p) and the X-specific part allowed us to distinguish X- and Y-bearing sperm (Fig. 4c). The distribution of Y1 is more similar to X1p than to X1q (Table 1). When we looked at the position of Y2 we found that this chromosome has a random distribution in sperm. All other sex chromosomes that we mapped are localized towards the posterior region of sperm (Table 3) with X and Y chromosomes showing very similar distributions. This is particularly obvious for Y4, X5, and Y5, which show equally high frequencies in the posterior of sperm. This suggests that the association of X and Y chromosomes is maintained beyond the formation of the meiotic chain in prophase I and that X and Y chromosomes occupy similar position in sperm (see “Discussion”). We determined a mostly non-random localization of platypus sex chromosomes, which share extensive homology with GGA Z. This shows that the arrangement of the mammalian homologs of GGA Z has changed to a non-random organization in early mammalian evolution.
Next, we localized a BAC clone from OAN 6 in sperm, which mapped very consistently to the middle of platypus sperm (see Fig. 4 and Table 3). The organization of this chromosome is similar to the distribution of the homologous GGA 4 (see “Discussion”).
Extensive differences in chromatin compaction are observed in chicken sperm
It is generally assumed that chromosomes are about six-fold more condensed in the sperm nucleus compared to the interphase nucleus (Haaf and Ward 1995; Mudrak et al. 2005). Elongated chromosome territories have been observed at different stages of spermatogenesis and in sperm in several species including mouse, rat, hamster, and pig (Foster et al. 2005; Meyer-Ficca et al. 1998; Ward et al. 1996). In chicken sperm, it was observed that the heterochromatic part of the Z chromosome showed a ten- to 15-fold extension compared to mitotic chromosomes (Solovei et al. 1998).
Here we wanted to investigate to what extent chromatin elongation is found in chicken sperm and if this is correlated to the total length of sperm nuclei or to random or non-random organization of the chromosome. We have selected BAC clones with known genomic distance on chicken chromosomes 1, 4, Z, and 27 to compare chromatin extension in MACs and MICs, in particular in MIC GGA 27, which organized non-randomly in chicken sperm. We compared the distance of the signals in metaphase chromosomes, interphase, and in sperm nuclei. The data clearly show that in elongated sperm the distance of clones is up to ten times larger than in metaphase and interphase chromatin (Table 4). As the distance between most chicken BAC clones is known from the chicken genome assembly, we could determine and compare the number of base pairs per micrometer length (Table 5). For example, the distance between the BAC clones 89F20 (23 Mb) and 66E20 (58 Mb) is 35 Mb on GGA Z. The mean distance between these two BAC clones in sperm is 4.5 μm or 7.8 Mb/μm. This is about ten-fold more than the values calculated for the Z heterochromatin (Solovei et al. 1998). On the other hand, the values obtained from euchromatic GGA 27 are only 0.4 Mb/μm.
In addition, we noted that the distance of the signals greatly varies between individual sperm. This ranges from co-localization to signals being situated at opposite ends of the sperm in both chicken and platypus (Figs. 3 and 4). Despite the seemingly random nature of chromatin elongation, the measurements correlated significantly with the total length of the sperm in most cases (Figs. 5, 6 and 7). Only the BAC clones for GGA 4 did not show a significant correlation between sperm length and distance of signals (data not shown).
Chromatin elongation of GGA 27 determined using FISH hybridization with BAC clones CH261-128H20 and CH261-18A6. One hundred twenty-seven hybridized chicken sperm were analyzed and measurements were plotted on a scatter plot graph (a). Descriptive statistics were generated for the data set (b). Despite a more than ten-fold difference in signal distance, regression analysis confirms the significant relationship between the two variables
Chromatin elongation of GGA Z determined using FISH hybridization with BAC clones CH261-66E20 and CH261-89F20. One hundred hybridized chicken sperm were analyzed and measurements were plotted on a scatter plot graph (a). Descriptive statistics were generated for the data set (b). Note a more than 20-fold difference in signal distance. Still regression analysis confirms a significant correlation between sperm length and signal distance
Chromatin elongation of OAN X1 determined using FISH hybridization with BAC clones CH236-286H10 and CH236-378F21. Fifty-eight hybridized sperm were analyzed and measurements were plotted on a scatter plot graph (a). Descriptive statistics were generated for the data set (b). The platypus X1 showed the highest fluctuation in signal distance (>30 times). Still a significant correlation was observed between distance of signals and sperm length
Chromosomes maintain a specific orientation in sperm
It was proposed that in sperm telomeres are attached to the nuclear membrane, and the proximity of chromosome arms in sperm suggests that chromosomes adopt a looped organization in sperm (Solov’eva et al. 2004). In elongated sperm, this would result in a chromosome maintaining a specific orientation within sperm even if the distribution of this chromosome is random along the sperm. In both chicken and platypus sperm, signal orientation was determined in a two-color FISH experiment with proximal and distal BAC clones from the acrocentric GGA 4 and OAN 6 and BAC clones from the long and short arms from GGA 27, GGA Z, and OAN X1. In each experiment, it was determined if the proximal or p arm signals were more anterior or more posterior than the distal or q arm signals (Table 6). The p values were calculated by chi-square to test against the null hypothesis that there is a random orientation of chromosomes within the sperm. We found that all chromosomes with the exception of GGA 27 maintained a specific orientation in sperm irrespective of random or non-random arrangement. GGA Z for example is randomly distributed in sperm but still maintains an orientation where the short arm points towards the posterior end and the long arm towards the anterior end of sperm. We were not able to determine an orientation for GGA 27. A possible explanation is that the close proximity of the two BAC clones (1.2 Mb) may be within hierarchical folding, which obscures detecting orientation in this case.
The first comparison of mammal and bird sperm reveals that the position of some chromosomes in sperm is highly conserved during evolution
Very few studies have investigated the evolutionary conservation of chromosome organization in mammalian sperm. Comparisons of chromosome positioning between species are difficult because of the variability of sperm shape in mammals. Here, we utilized the fact that platypus and chicken sperm are very similar in shape and size, which allows precise comparison of chromosome positioning between both species. We investigated the position of GGA 1 and 4, which share large blocks of homology with the mammalian X. GGA 4 shows a non-random organization in the middle of the sperm nucleus, while GGA 1 is randomly organized. Interestingly, OAN 6, which shares extensive homology with the mammalian X (and GGA 4), is also found in the medial part of platypus sperm demonstrating a high evolutionary conservation of the position for these homologous chromosomes.
The MHC is autosomal in humans (chromosome 6) and chicken (GGA 17) but on sex chromosomes in platypus (Y3X3 and Y4X5) and echidna (Dohm et al. 2007). In human sperm, chromosome 6 is located in the medial part of sperm towards the nuclear periphery (Zalenskaya and Zalensky 2004). We determined that GGA 17 is organized non-randomly in the middle of sperm as well. In platypus, the MHC is disrupted and clusters with most of the other sex chromosomes in the posterior of sperm. Interestingly, in hamster sperm a MHC class I gene has been mapped to the posterior part of sperm as well (Ward et al. 1996).
GGA 27 is arranged non-randomly in the middle of chicken sperm. GGA 27 shares extensive homology with human chromosome 17, which also localizes in the middle of human sperm (Zalenskaya and Zalensky 2004). This is the first time that chromosome positioning has been compared between chicken and mammals, which reveals that the position of some chromosomes has been conserved over several hundred years of divergent evolution (see “Discussion”).
Discussion
The advent of whole-genome sequencing efforts provides a huge range of new tools and DNA probes that enable us to study genome organization of specific regions in sperm. Available sequence of these probes also allows sequence-based comparisons between species. In this study, we have utilized BAC clones available from the chicken and platypus whole-genome sequencing projects as well as a set of sex chromosome-specific BAC clones from platypus that we have generated.
Recent studies of genome organization in birds and mammals have proposed that genome organization in sperm is very different in both groups (Greaves et al. 2003; Solovei et al. 1998). In mammals, generally, a higher-order organization is found with most chromosomes occupying defined positions in sperm. It has been suggested that chromosome size, sperm shape, and specific function as centromere, telomeres, or sex chromosome are factors influencing chromosome positioning in sperm (Meyer-Ficca et al. 1998; Solov’eva et al. 2004; Solovei et al. 1998). However, a thorough study in pig spermatozoa found no evidence that smaller chromosome show a different organization compared to larger chromosomes (Foster et al. 2005) and our data support this. We have included large and small chromosomes in both species and find no evidence that small chromosomes are more likely to be organized at random in sperm.
Higher-order genome organization is present in chicken sperm
Generally, in chicken, the random distribution of chromosomes and extensive chromatin elongation were interpreted as a general lack of higher-order organization (Greaves et al. 2003; Solovei et al. 1998). One of the main outcomes of the present study is that we have observed a higher-order organization of chromosomes in chicken sperm: we found that the location of five of the 12 chromosomes tested was not random (Table 2) as determined by a significant deviation from an expected random distribution of signals with frequencies of 33% in any region of the chicken sperm. The highest frequencies did not exceed 56% for any clone (Table 1). This is a lower frequency than that observed for most mammalian chromosomes (>70% in many cases) with the exception of human chromosome 13, which is randomly distributed (Hazzouri et al. 2000). It is difficult at this stage to speculate about the functional relevance of this gradual difference; however, it is clear that the general view of a random organization in chicken sperm as opposed to a non-random organization in mammalian sperm needs to be revised. Platypus and chicken sperm have similar sperm shape and both lack bisulfite bonds of protamine molecules. In addition, they share vast size differences of chromosomes within their karyotypes. Despite all these reptilian features of platypus sperm and chromosomes, we observed that chromosomes often occupy a certain position in well above 60% of the nuclei in platypus (see Table 5), which is similar to other mammals. In chicken sperm, these frequencies were lower and we never observed more than 56% of the signals in any particular part of the sperm nucleus. We conclude that the lower frequencies observed in chicken sperm must be the result of factors other than sperm shape protamine compaction and chromosome size. Interestingly, we observed that in particular in chicken pairs of BAC clones from the same chromosomes showed different distribution (e.g., GGA 1, GGA 4, GGA Z). In these cases, some clones show a random distribution, while other clones from the same chromosome show a more specific non-random localization. In a loop-like organization, it may be plausible that clones with a more telomeric position show a more defined position than clones from regions further out on the loops. However, our data indicate that distal or proximal position of the BAC clone does not correlate with non-random or random positioning, respectively (Table 1).
Extensive differences in chromatin compaction are observed in chicken sperm
In mammals, sperm chromatin is four to six times more compact than that of metaphase chromosome (Balhorn 1982; Haaf and Ward 1995; Mudrak et al. 2005). In situ hybridization approaches using rDNA, repetitive elements, and chromosome paints have been used on mildly decondensed human and mouse sperm chromatin. In mouse, rod-like elongated signals were observed and support a linear array model of sperm chromatin (Haaf and Ward 1995). Recent work using chromosome arm-specific painting probes showed a very compact chromosome territory structure in human sperm (Mudrak et al. 2005).
In chicken, a probe of the heterochromatic block on the Z chromosome showed a characteristic dynamic from very compact in early stages of spermatogenesis to very elongated in the spermatid nucleus (Solovei et al. 1998). Similar stretched out chromatin has been observed in elongated sperm in other vertebrate and non-vertebrates species (Joffe et al. 1998). One of the advantages of using well-defined BAC clones is that the exact genomic distance is known for clones that have been mapped to whole-genome assemblies. As this was the case for the chicken BAC clones, we could calculate and quantify the extent of chromatin compaction. A previous study used a probe of the heterochromatic part of the Z chromosome and estimated that this region is about ten to 15 times longer in sperm compared to mitotic chromosomes and would contain about 0.7 Mb/μm (Solovei et al. 1998). We found massive differences in both distance of signals and compaction of chromatin (measured in Mb/μm). For example, we determined 400 kb/μm for BAC clones only 1.2 Mb apart on GGA 27 and 7.8 Mb/μm for BAC clones spanning 35 Mb including the GGA Z centromere. The ten-fold difference of chromatin compaction between the heterochromatic region on Zp reported by Solovei et al. (1998) and the pericentromeric region in this present study indicate that centromeres are extremely condensed in sperm. The formation of a compact chromocenter has been reported in mouse, rat and human and chicken (Brinkley 1986; Dressler and Schmid 1976; Meyer-Ficca et al. 1998; Zalensky et al. 1993). Interestingly, in chicken, it was reported that centromeric heterochromatin is organized non-randomly in sperm and adopts a tandem organization across sperm (Dressler and Schmid 1976). The random organization of the pericentromeric BAC clones from GGA Z used in this study indicates a more random organization of centromeres, but the much higher condensation of the centromeric region supports the idea that a compact chromocenter exists in birds. This compaction difference may also be explained by a different organization of chromatin in the sperm nucleus. For the heterochromatic region on GGA Z, reorganization during spermatogenesis was observed leading to a stretched out pattern in sperm reminiscent of a spiral-type arrangement observed with 28S rDNA in the elongated sperm of planarians (Joffe et al. 1998), indicating that chromatin fibers in elongated sperm could be stacked.
Our data show that almost 20-fold differences in compaction are present in sperm despite the replacement of histones by protamines, following the general observation of less condensed euchromatin (e.g., the gene-rich GGA 27) and more condensed heterochromatin (e.g., the GGA Z centromere) in metaphase and interphase chromatin. Generally, sperm chromatin was significantly more elongated compared to metaphase or interphase chromatin (Table 5). It would be interesting to investigate whether similar regional differences are also observed in eutherian sperm. In platypus, we also observed chromatin extension for X1 and OAN 6 in sperm. However, as the physical distances between these clones is not known, a direct comparison with chicken is not possible.
The distance of clones within sperm correlates with total sperm length
We have used BAC clones anchored in the chicken genome and found a significant correlation between the distance of signals and overall length of the sperm nucleus in both species (Figs. 5, 6 and 7). This was found independently of random or non-random distribution of BAC clones in chicken. The only chromosome where this correlation was not observed is the acrocentric GGA 4. The large distance of clones within sperm is quite different to the organization in human sperm, where proximity of telomeric and subtelomeric regions supports the idea of a looped structure in the sperm nucleus (Solov’eva et al. 2004). Although the probes used in this study were not subtelomeric, the much larger distance of the clones suggests that in elongated sperm the proximity of chromosome arms has been lost most likely through the extreme elongation of the sperm nucleus in platypus and chicken.
Chromosomes maintain a specific orientation in sperm
Another indication of higher-order organization in chicken sperm is our observation, that chromosomes maintain a specific orientation in sperm. The use of chromosome-specific BAC clones allowed us to determine the orientation of a chromosome in sperm. We found that GGA Z (randomly organized) and GGA 4 (non-randomly organized) maintain a specific orientation in sperm. We could not determine a specific direction for GGA 27, possibly due to the close proximity of the BAC clones that were used as probes. In platypus, we investigated orientation of BAC clones on OAN 6 and on the short arm of OAN X1. For both chromosomes, we found that they maintain a specific direction (Table 6). This indicates that higher-order organization of the genome may also include that chromosomes are organized in a certain direction within the sperm nucleus.
The platypus sperm nucleus is elongated and very similar compared to chicken sperm. A thorough comparison of quail and platypus spermatogenesis has revealed many similarities and some differences between both species (Lin and Jones 2000). A similar picture emerges in terms of genome organization in sperm. Generally, platypus chromosomes are non-randomly organized within the sperm nucleus as observed for other mammals. The comparison with chicken revealed similarities between genome organization in platypus and chicken sperm: in both species the distance of the signals correlate with the sperm length (Figs. 5, 6 and 7). This could be a common feature of elongated sperm and seems independent of organization (random or non-random) or chromosome size. In addition, the platypus features a unique clustering of sex chromosomes in sperm.
Non-random organization of autosomes and clustering of multiple sex chromosomes are present in platypus sperm
To investigate whether platypus sex chromosomes remain associated after they segregated from the sex chromosome chain that links all ten sex chromosomes during meiotic prophase I in males (Grutzner et al. 2004), we mapped BAC clones from seven platypus sex chromosomes in sperm. We have mapped X1 to the medial region of sperm. However, when we look at the localization of the X1p and X1q BAC clones separately, our results show almost equal numbers of signals in the anterior and the middle of sperm for X1p. In contrast, the long arm is found more often in the middle. This shows that X1 retains an orientation where the short arm points towards the anterior of the sperm nucleus (Table 6). Interestingly, Y1 shares extensive homology with X1p and shows a very similar distribution. Y2 shows a random organization in sperm. Y3, Y4, X5, and Y5 are all located in the posterior region of sperm. This shows very similar distribution of X and Y chromosomes in platypus sperm. Proximity of X and Y chromosomes has also been observed in pigs (Foster et al. 2005). More importantly, a clustering of most platypus sex chromosomes in platypus sperm is evident and suggests that the multiple sex chromosomes in platypus maintain association after the meiotic chain has segregated into X1–X5 and Y1–Y5 bearing secondary spermatocytes. If this is the result of meiotic or postmeiotic silencing in monotremes is currently unknown.
The position of some chromosomes in sperm is highly conserved during amniote evolution
The use of genomic clones from sequenced and assembled genomes offers a unique opportunity for comparing the dynamics of chromosome positioning during evolution. For example, HSA 17 is one of the most gene-rich chromosomes in humans and occupies a central position in the medial part of the elliptic human sperm (Hazzouri et al. 2000). Interestingly, GGA 27, which shares homology to HSA 17, also locates in the middle of sperm. The conservation of chromosome position within sperm over long evolutionary distances highlights the functional importance of genome organization in sperm.
The position of sex chromosomes in sperm is particularly interesting as epigenetic changes leading meiotic sex chromosome inactivation during spermatogenesis could affect positioning of sex chromosomes in sperm. In mammals, sex chromosomes are silenced during meiosis and the X and Y chromosomes reside as inactive chromatin in a separate compartment called the “sex body” (Handel 2004; Turner 2007). It has recently been shown that meiotic sex chromosome inactivation is maintained beyond the first meiotic division as postmeiotic sex chromatin in mouse (Namekawa et al. 2006) and marsupials (Namekawa et al. 2007). It is currently unknown if sex chromosome silencing occurs in female chicken or in male platypus.
It was also proposed that positioning of the X chromosome in sperm could play a role in the preferential inactivation of the paternal X chromosome in the early embryo. Recently, research revealed a regional pattern of dosage compensation in birds (Melamed and Arnold 2007), and emerging evidence suggests a similar pattern of dosage compensation in platypus (Deakin et al. 2008).
Most of the genes from chicken Z are found on human chromosome 9, while most of the genes of the therian X are found on chicken chromosomes 1 and 4. Platypus shares an XY sex chromosomes system with other mammals but its sex chromosomes show extensive homology to the chicken Z (Grutzner et al. 2004; Veyrunes et al. 2008). One of the exceptions to this is MHC genes that have recently mapped to the pseudoautosomal regions of two pairs of platypus sex chromosomes (Y3X3 and Y4X5; Dohm et al. 2007). In human and chicken, the MHC is autosomal (HSA 6 and GGA 16 and 17). Most genes from the human X reside on OAN 6 (Veyrunes et al. 2008; Waters et al. 2005). To shed some light on the evolutionary dynamic of sex chromosomes and autosomes with homology to sex chromosomes in birds and mammals, we mapped BAC clones from GGA 1, 4 and GGA Z, GGA 17, OAN 6 and most platypus sex chromosomes in platypus and chicken sperm.
Our results confirm a random organization of the chicken Z chromosome in sperm as reported previously (Solovei et al. 1998). Platypus sex chromosomes with homology to the chicken Z are mostly found in the posterior, which shows that these chromosomes have changed from a random pattern in chicken to a non-random distribution in platypus sperm. In addition, MHC genes on GGA 17 have moved from a medial position in chicken to posterior in platypus.
Most of the genes from human Xp are found on chicken chromosome 1, while most of the genes from human Xq are clustered on the short arm of GGA 4 (Kohn et al. 2007). We found a random arrangement of BAC clones from chromosome 1, but a non-random location of GGA 4. The BAC clones for GGA 4 are from the region homologous to human Xq (Kohn et al. 2007). ONA 6, which shares extensive homology with the human X, also resides in the middle of the elongated platypus sperm nucleus. This position has therefore been conserved in birds and monotreme mammals and is also found in some therian mammals. In contrast, in several other eutherian and marsupial species the X can be found in the anterior part of sperm (Foster et al. 2005; Greaves et al. 2003; Hazzouri et al. 2000). We conclude that repositioning of sex chromosomes to the anterior part of sperm is derived and has occurred in several mammalian lineages independently (Fig. 8).
The evolution of sex chromosomes organization in amniote sperm. This study indicates that higher-order organization is found not only in mammals but also reptiles. On top, homology relationships of sex chromosomes systems and dosage compensation mechanisms are shown. In monotremes, first data indicate patchy dosage compensation. There have been three major changes of sex chromosome organization in amniotes: a shift of random organization of GGA Z towards non-random organization in platypus. The anterior position in sperm of most eutherian mammals indicates that the X could be the first chromosome to make contact to the oocyte, which is not the case for the autosomal homologs in chicken and platypus
Conclusion
This study is the most extensive study of genome organization in the elongated chicken and platypus sperm and contributes new evidence to the growing body of data that shows the importance of genome organization in sperm in various species. We provide first evidence for chromosome specific higher-order organization in the chicken sperm nucleus. In platypus, our findings show a clustering of most sex chromosomes in the posterior part of sperm, indicative of postmeiotic association of X and Y chromosomes.
Localization of platypus and chicken sex chromosomes and autosomes that share homology to the therian X in sperm suggests that the chicken Z chromosome has adopted a non-random organization in early mammalian evolution. The autosomes homologous to the therian X have maintained their position in the middle of the sperm nucleus until in some lineages they moved towards the anterior.
In addition, this study shows how well-defined genomic clones can be used to investigate new aspects of genome organization in the sperm nucleus. For example, we show that orientation of chromosomes is maintained in sperm. We also discovered that large differences in chromatin compaction exists within and between chromosomes, suggesting that chromatin condensation in the sperm nucleus follows the same pattern as in metaphase or interphase chromatin.
References
Balhorn R (1982) A model for the structure of chromatin in mammalian sperm. J Cell Biol 93:298–305
Bedford JM (1991) The coevolution of mammalian gametes. In: Dunbar BS, O’Rand MG (eds) A comparative overview of mammalian fertilization. Plenum, New York, pp 3–35
Bedford JM (2004) Enigmas of mammalian gamete form and function. Biol Rev Camb Philos Soc 79:429–460
Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A (2007) The delayed rise of present-day mammals. Nature 446:507–512
Brinkley BR (1986) Arrangements of kinetochores in mouse cells during meiosis and spermiogenesis. Chromosoma 94:266–272
Cremer M, von Hase J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T (2001) Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res 9:541–567
Deakin JE, Hore TA, Koina E, Marshall Graves JA (2008) The status of dosage compensation in the multiple X chromosomes of the platypus. PLoS Genet 4(7):e1000140
Dohm JC, Tsend-Ayush E, Reinhardt R, Grutzner F, Himmelbauer H (2007) Disruption and pseudoautosomal localization of the major histocompatibility complex in monotremes. Genome Biol 8:R175
Dressler B, Schmid M (1976) Specific arrangement of chromosomes in the spermiogenesis of Gallus domesticus. Chromosoma 58:387–391
Fillon V, Morisson M, Zoorob R, Auffray C, Douaire M, Gellin J, Vignal A (1998) Identification of 16 chicken microchromosomes by molecular markers using two-colour fluorescence in situ hybridization (FISH). Chromosome Res 6:307–313
Foster HA, Abeydeera LR, Griffin DK, Bridger JM (2005) Non-random chromosome positioning in mammalian sperm nuclei, with migration of the sex chromosomes during late spermatogenesis. J Cell Sci 118:1811–1820
Fraser P, Bickmore W (2007) Nuclear organization of the genome and the potential for gene regulation. Nature 447:413–417
Greaves IK, Svartman M, Wakefield M, Taggart D, De Leo A, Ferguson-Smith MA, Rens W, O’Brien PC, Voullaire L, Westerman M, Graves JA (2001) Chromosomal painting detects non-random chromosome arrangement in dasyurid marsupial sperm. Chromosome Res 9:251–259
Greaves IK, Rens W, Ferguson-Smith MA, Griffin D, Marshall Graves JA (2003) Conservation of chromosome arrangement and position of the X in mammalian sperm suggests functional significance. Chromosome Res 11:503–512
Grutzner F, Rens W, Tsend-Ayush E, El-Mogharbel N, O’Brien PC, Jones RC, Ferguson-Smith MA, Marshall Graves JA (2004) In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature 432:913–917
Haaf T, Ward DC (1995) Higher order nuclear structure in mammalian sperm revealed by in situ hybridization and extended chromatin fibers. Exp Cell Res 219:604–611
Habermann FA, Cremer M, Walter J, Kreth G, von Hase J, Bauer K, Wienberg J, Cremer C, Cremer T, Solovei I (2001) Arrangements of macro- and microchromosomes in chicken cells. Chromosome Res 9:569–584
Handel MA (2004) The XY body: a specialized meiotic chromatin domain. Exp Cell Res 296:57–63
Hazzouri M, Rousseaux S, Mongelard F, Usson Y, Pelletier R, Faure AK, Vourc’h C, Sele B (2000) Genome organization in the human sperm nucleus studied by FISH and confocal microscopy. Mol Reprod Dev 55:307–315
Heard E, Disteche CM (2006) Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20:1848–1867
Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Dodgson JB (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716
Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CCJ, Wright MW, Wain HM, Ziegler A, Beck S (2004) Gene map of the extended human MHC. Nat Rev Genet 12:889–899
Joffe BI, Solovei IV, Macgregor HC (1998) Ordered arrangement and rearrangement of chromosomes during spermatogenesis in two species of planarians (Plathelminthes). Chromosoma 107:173–183
Kimmins S, Sassone-Corsi P (2005) Chromatin remodelling and epigenetic features of germ cells. Nature 434:583–589
Kohn M, Kehrer-Sawatzki H, Steinbach P, Marshall Graves JA, Hameister H (2007) Recruitment of old genes to new functions: evidences obtained by comparing the orthologues of human XLMR genes in mouse and chicken. Cytogenet Genome Res 116:173–180
Leung LK, Cummins JM (1988) Morphology of immature spermatozoa of the Chinese pangolin (Manis pentadactyla: Pholidota). Proceedings of the Australian Society of Reproductive Biology 20
Lin M, Jones RC (2000) Spermiogenesis and spermiation in a monotreme mammal, the platypus, Ornithorhynchus anatinus. J Anat 196(Pt 2):217–232
Matthey R (1949) Les chromosomes des vertebrees. F. Rouge, Lucerne
McQueen HA, Fantes J, Cross SH, Clark VH, Archibald AL, Bird AP (1996) CpG islands of chicken are concentrated on microchromosomes. Nat Genet 12:321–324
McQueen HA, Siriaco G, Bird AP (1998) Chicken microchromosomes are hyperacetylated, early replicating, and gene rich. Genome Res 8:621–630
Melamed E, Arnold AP (2007) Regional differences in dosage compensation on the chicken Z chromosome. Genome Biol 8:R202
Meyer-Ficca M, Muller-Navia J, Scherthan H (1998) Clustering of pericentromeres initiates in step 9 of spermiogenesis of the rat (Rattus norvegicus) and contributes to a well defined genome architecture in the sperm nucleus. J Cell Sci 111(Pt 10):1363–1370
Mudrak O, Tomilin N, Zalensky A (2005) Chromosome architecture in the decondensing human sperm nucleus. J Cell Sci 118:4541–4550
Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT (2006) Postmeiotic sex chromatin in the male germline of mice. Curr Biol 16:660–667
Namekawa SH, VandeBerg JL, McCarrey JR, Lee JT (2007) Sex chromosome silencing in the marsupial male germ line. Proc Natl Acad Sci U S A 104:9730–9735
Nanda I, Schmid M (1994) Localization of the telomeric (TTAGGG)n sequence in chicken (Gallus domesticus) chromosomes. Cytogenet Cell Genet 65:190–193
Nanda I, Shan Z, Schartl M, Burt DW, Koehler M, Nothwang H, Grützner F, Paton IR, Windsor D, Dunn I, Engel W, Staeheli P, Mizuno S, Haaf T, Schmid M (1999) 300 million years of conserved synteny between chicken Z and human chromosome 9. Nat Genet 3:258–259
Oliva R, Dixon GH (1989) Chicken protamine genes are intronless. The complete genomic sequence and organization of the two loci. J Biol Chem 264:12472–12481
Rens W, Grutzner F, O’Brien PC, Fairclough H, Graves JA, Ferguson-Smith MA (2004) Resolution and evolution of the duck-billed platypus karyotype with an X1Y1X2Y2X3Y3X4Y4X5Y5 male sex chromosome constitution. Proc Natl Acad Sci U S A 101:16257–16261
Retief JD, Winkfein RJ, Dixon GH (1993) Evolution of the monotremes. The sequences of the protamine P1 genes of platypus and echidna. Eur J Biochem 218:457–461
Solov’eva L, Svetlova M, Bodinski D, Zalensky AO (2004) Nature of telomere dimers and chromosome looping in human spermatozoa. Chromosome Res 12:817–823
Solovei IV, Joffe BI, Hori T, Thomson P, Mizuno S, Macgregor HC (1998) Unordered arrangement of chromosomes in the nuclei of chicken spermatozoa. Chromosoma 107:184–188
Turner JM (2007) Meiotic sex chromosome inactivation. Development 134:1823–1831
Van Brink J (1959) L’ expression morphologique de la diagametie chez les sauropsids et les monotremes. Chromosoma 10:1–72
Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, Grutzner F, Deakin JE, Whittington CM, Schatzkamer K, Kremitzki CL, Graves T, Ferguson-Smith MA, Warren W, Marshall Graves JA (2008) Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res 18:965–973
Wallis JW, Aerts J, Groenen MA, Crooijmans RP, Layman D, Graves TA, Scheer DE, Kremitzki C, Fedele MJ, Mudd NK, Cardenas M, Higginbotham J, Carter J, McGrane R, Gaige T, Mead K, Walker J, Albracht D, Davito J, Yang SP, Leong S, Chinwalla A, Sekhon M, Wylie K, Dodgson J, Romanov MN, Cheng H, de Jong PJ, Osoegawa K, Nefedov M, Zhang H, McPherson JD, Krzywinski M, Schein J, Hillier L, Mardis ER, Wilson RK, Warren WC (2004) A physical map of the chicken genome. Nature 432:761–764
Ward WS, McNeil J, de Lara J, Lawrence J (1996) Localization of three genes in the hook-shaped hamster sperm nucleus by fluorescent in situ hybridization. Biol Reprod 54:1271–1278
Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grutzner F, Belov K, Miller W, Clarke L, Chinwalla AT, Yang SP, Heger A, Locke DP, Miethke P, Waters PD, Veyrunes F, Fulton L, Fulton B, Graves T, Wallis J, Puente XS, Lopez-Otin C, Ordonez GR, Eichler EE, Chen L, Cheng Z, Deakin JE, Alsop A, Thompson K, Kirby P, Papenfuss AT, Wakefield MJ, Olender T, Lancet D, Huttley GA, Smit AF, Pask A, Temple-Smith P, Batzer MA, Walker JA, Konkel MK, Harris RS, Whittington CM, Wong ES, Gemmell NJ, Buschiazzo E, Vargas Jentzsch IM, Merkel A, Schmitz J, Zemann A, Churakov G, Kriegs JO, Brosius J, Murchison EP, Sachidanandam R, Smith C, Hannon GJ, Tsend-Ayush E, McMillan D, Attenborough R, Rens W, Ferguson-Smith M, Lefevre CM, Sharp JA, Nicholas KR, Ray DA, Kube M, Reinhardt R, Pringle TH, Taylor J, Jones RC, Nixon B, Dacheux JL, Niwa H, Sekita Y, Huang X, Stark A, Kheradpour P, Kellis M, Flicek P, Chen Y, Webber C, Hardison R, Nelson J, Hallsworth-Pepin K, Delehaunty K, Markovic C, Minx P, Feng Y, Kremitzki C, Mitreva M, Glasscock J, Wylie T, Wohldmann P, Thiru P, Nhan MN, Pohl CS, Smith SM, Hou S, Renfree MB, Mardis ER, Wilson RK (2008) Genome analysis of the platypus reveals unique signatures of evolution. Nature 453:175–183
Waters PD, Delbridge ML, Deakin JE, El-Mogharbel N, Kirby PJ, Carvalho-Silva DR, Graves JA (2005) Autosomal location of genes from the conserved mammalian X in the platypus (Ornithorhynchus anatinus): implications for mammalian sex chromosome evolution. Chromosome Res 13:401–410
Watson JM, Meyne J, Graves JA (1996) Ordered tandem arrangement of chromosomes in the sperm heads of monotreme mammals. Proc Natl Acad Sci U S A 93:10200–10205
White MJD (1973) Animal cytology and evolution. Cambridge University Press, London
Zalenskaya IA, Zalensky AO (2004) Non-random positioning of chromosomes in human sperm nuclei. Chromosome Res 12:163–173
Zalensky AO, Breneman JW, Zalenskaya IA, Brinkley BR, Bradbury EM (1993) Organization of centromeres in the decondensed nuclei of mature human sperm. Chromosoma 102:509–518
Acknowledgments
We are grateful to Megan Wright for excellent technical assistance and Felicia Paulraj for contributing data on chicken chromosome 16 and Prof. Russell Jones for support with the monotreme sample collection. We thank Glenrock Station and Macquarie Generation for providing facilities; Prof. Bill Breed for critically reading the manuscript; and Dr. Jack daSilva, Dr. Daniel Kortschak, and Prof. David Adelson for their help with the statistical analysis. The work was supported by the Australian Research Council Discovery Project and Fellowship schemes. Frank Grützner is an Australian Research Council Australian Research Fellow and Enkhjargal Tsend-Ayush is an Australian Research Council Postdoctoral Fellow.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E.A. Nigg
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Tsend-Ayush, E., Dodge, N., Mohr, J. et al. Higher-order genome organization in platypus and chicken sperm and repositioning of sex chromosomes during mammalian evolution. Chromosoma 118, 53–69 (2009). https://doi.org/10.1007/s00412-008-0177-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-008-0177-1