Abstract
Pituitary blastoma (PitB) has recently been identified as a rare and potentially lethal pediatric intracranial tumor. All cases that have been studied molecularly possess at least one DICER1 pathogenic variant. Here, we characterized nine pituitary samples, including three fresh frozen PitBs, three normal fetal pituitary glands and three normal postnatal pituitary glands using small-RNA-Seq, RNA-Seq, methylation profiling, whole genome sequencing and Nanostring® miRNA analyses; an extended series of 21 pituitary samples was used for validation purposes. These analyses demonstrated that DICER1 RNase IIIb hotspot mutations in PitBs induced improper processing of miRNA precursors, resulting in aberrant 5p-derived miRNA products and a skewed distribution of miRNAs favoring mature 3p over 5p miRNAs. This led to dysregulation of hundreds of 5p and 3p miRNAs and concomitant dysregulation of numerous mRNA targets. Gene expression analysis revealed PRAME as the most significantly upregulated gene (500-fold increase). PRAME is a member of the Retinoic Acid Receptor (RAR) signaling pathway and in PitBs, the RAR, WNT and NOTCH pathways are dysregulated. Cancer Hallmarks analysis showed that PI3K pathway is activated in the tumors. Whole genome sequencing demonstrated a quiet genome with very few somatic alterations. The comparison of methylation profiles to publicly available data from ~ 3000 other central nervous system tumors revealed that PitBs have a distinct methylation profile compared to all other tumors, including pituitary adenomas. In conclusion, this comprehensive characterization of DICER1-related PitB revealed key molecular underpinnings of PitB and identified pathways that could potentially be exploited in the treatment of this tumor.




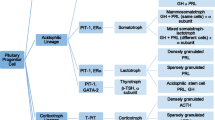
adapted from ref [23]. Consistent with activation of WNT and NOTCH pathways in PitB and normal fetal pituitary (NFP) samples, these samples show higher level of undifferentiated cells compared to normal postnatal pituitary (NPP) samples. PitB samples have lower level of differentiated cells compared to both NPP and NFP samples (see Online Resource 11 for a full list of the genes which were selected from the list of mouse genes listed in ref [10])



Similar content being viewed by others
References
Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. https://doi.org/10.1093/bioinformatics/btu638
Andrews S, FastQC (2019) A Quality Control tool for High Throughput Sequence Data. http://www.bioinformaticsbabrahamacuk/projects/fastqc/: Doi citeulike-article-id:11583827
Anglesio M, Wang Y, Yang W, Senz J, Wan A, Heravi-Moussavi A et al (2012) Cancer-associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J Pathol 229:400–409. https://doi.org/10.1002/path.4135
Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD et al (2014) Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30:1363–1369. https://doi.org/10.1093/bioinformatics/btu049
Boeva V, Popova T, Bleakley K, Chiche P, Cappo J, Schleiermacher G et al (2012) Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28:423–425. https://doi.org/10.1093/bioinformatics/btr670
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Boon K, Edwards JB, Siu IM, Olschner D, Eberhart CG, Marra MA et al (2003) Comparison of medulloblastoma and normal neural transcriptomes identifies a restricted set of activated genes. Oncogene 22:7687–7694. https://doi.org/10.1038/sj.onc.1207043
Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D et al (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–474. https://doi.org/10.1038/nature26000
Chen J, Wang Y, McMonechy MK, Anglesio MS, Yang W, Senz J et al (2015) Recurrent DICER1 hotspot mutations in endometrial tumours and their impact on microRNA biogenesis. J Pathol 237:215–225. https://doi.org/10.1002/path.4569
Cheung LYM, George AS, McGee SR, Daly AZ, Brinkmeier ML, Ellsworth BS et al (2018) Single-cell RNA sequencing reveals novel markers of male pituitary stem cells and hormone-producing cell types. Endocrinology 159:3910–3924. https://doi.org/10.1210/en.2018-00750
Davis SW, Castinetti F, Carvalho LR, Ellsworth BS, Potok MA, Lyons RH et al (2010) Molecular mechanisms of pituitary organogenesis: In search of novel regulatory genes. Mol Cell Endocrinol 323:4–19. https://doi.org/10.1016/j.mce.2009.12.012
de Kock L, Priest JR, Foulkes WD, Alexandrescu S (2020) An update on the central nervous system manifestations of DICER1 syndrome. Acta Neuropathol 139:689–701. https://doi.org/10.1007/s00401-019-01997-y
de Kock L, Sabbaghian N, Plourde F, Srivastava A, Weber E, Bouron-Dal Soglio D et al (2014) Pituitary blastoma: a pathognomonic feature of germ-line DICER1 mutations. Acta Neuropathol 128:111–122. https://doi.org/10.1007/s00401-014-1285-z
de Kock L, Wu MK, Foulkes WD (2019) Ten years of DICER1 mutations: Provenance, distribution, and associated phenotypes. Hum Mutat 40:1939–1953. https://doi.org/10.1002/humu.23877
Devnath S, Inoue K (2008) An insight to pituitary folliculo-stellate cells. J Neuroendocrinol 20:687–691. https://doi.org/10.1111/j.1365-2826.2008.01716.x
Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R (2005) The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell 122:835–847. https://doi.org/10.1016/j.cell.2005.07.003
Foulkes WD, Priest JR, Duchaine TF (2014) DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer 14:662–672. https://doi.org/10.1038/nrc3802
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. https://doi.org/10.1186/gb-2004-5-10-r80
Guaraldi F, Storr HL, Ghizzoni L, Ghigo E, Savage MO (2014) Paediatric pituitary adenomas: a decade of change. Horm Res Paediatr 81:145–155. https://doi.org/10.1159/000357673
Guillerman RP, Foulkes WD, Priest JR (2019) Imaging of DICER1 syndrome. Pediatr Radiol 49:1488–1505. https://doi.org/10.1007/s00247-019-04429-x
Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, Prentice L et al (2012) Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med 366:234–242. https://doi.org/10.1056/NEJMoa1102903
Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D et al (2009) DICER1 mutations in familial pleuropulmonary blastoma. Science 325:965. https://doi.org/10.1126/science.1174334
Ho Y, Hu P, Peel MT, Chen S, Camara PG, Epstein DJ et al (2019) Single cell transcriptomic analysis of the adult mouse pituitary reveals a novel multi-hormone cell cluster and physiologic demand-induced lineage plasticity. Biorxiv 2019:475558. https://doi.org/10.1101/475558
Horvath E, Coire CI, Kovacs K, Smyth HS (2010) Folliculo-stellate cells of the human pituitary as adult stem cells: examples of their neoplastic potential. Ultrastruct Pathol 34:133–139. https://doi.org/10.3109/01913121003662247
Horvath E, Kovacs K (2002) Folliculo-stellate cells of the human pituitary: a type of adult stem cell? Ultrastruct Pathol 26:219–228. https://doi.org/10.1080/01913120290104476
Ikeda H, Lethe B, Lehmann F, van Baren N, Baurain JF, de Smet C et al (1997) Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity 6:199–208. https://doi.org/10.1016/s1074-7613(00)80426-4
Jessa S, Blanchet-Cohen A, Krug B, Vladoiu M, Coutelier M, Faury D et al (2019) Stalled developmental programs at the root of pediatric brain tumors. Nat Genet 51:1702–1713. https://doi.org/10.1038/s41588-019-0531-7
Keil MF, Stratakis CA (2008) Pituitary tumors in childhood: update of diagnosis, treatment and molecular genetics. Expert Rev Neurother 8:563–574. https://doi.org/10.1586/14737175.8.4.563
Kelberman D, Rizzoti K, Lovell-Badge R, Robinson IC, Dattani MT (2009) Genetic regulation of pituitary gland development in human and mouse. Endocr Rev 30:790–829. https://doi.org/10.1210/er.2009-0008
Khan NE, Bauer AJ, Schultz KAP, Doros L, Decastro RM, Ling A et al (2017) Quantification of thyroid cancer and multinodular Goiter risk in the DICER1 syndrome: a family-based cohort study. J Clin Endocrinol Metab 102:1614–1622. https://doi.org/10.1210/jc.2016-2954
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. https://doi.org/10.1186/gb-2009-10-3-r25
Larkin S, Ansorge O (2000) Development and microscopic anatomy of the pituitary gland. In: Feingold KR, Anawalt B, Boyce A, Chrousos G, de Herder WW, Dungan K, Grossman A, Hershman JM, Hofland J, Kaltsas Get al (eds) Endotext. MDText.com, Inc
Law CW, Chen Y, Shi W, Smyth GK (2014) voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29. https://doi.org/10.1186/gb-2014-15-2-r29
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/bioinformatics/btt656
Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P (2015) The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 1:417–425. https://doi.org/10.1016/j.cels.2015.12.004
Lin S, Gregory RI (2015) MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15:321–333. https://doi.org/10.1038/nrc3932
Liu APY, Kelsey MM, Sabbaghian N, Park SH, Deal CL, Esbenshade AJ et al (2020) Clinical outcomes and complications of pituitary blastoma. J Clin Endocrinol Metab 106:351–363. https://doi.org/10.1210/clinem/dgaa857
Lopes MBS (2017) The 2017 World Health Organization classification of tumors of the pituitary gland: a summary. Acta Neuropathol 134:521–535. https://doi.org/10.1007/s00401-017-1769-8
Mannelli M, Canu L, Ercolino T, Rapizzi E, Martinelli S, Parenti G et al (2018) Diagnosis of endocrine disease: SDHx mutations: beyond pheochromocytomas and paragangliomas. Eur J Endocrinol 178:R11–R17. https://doi.org/10.1530/EJE-17-0523
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A et al (2016) The ensembl variant effect predictor. Genome Biol 17:122. https://doi.org/10.1186/s13059-016-0974-4
Mello BP, de Carvalho DD, Campos AH, Soares FA, Amarante-Mendes GP (2013) Regulation of TRAIL expression by PRAME and EZH2 as potential therapeutic target against solid tumors. BMC Proc 7:P10. https://doi.org/10.1186/1753-6561-7-s2-p10
Moore KL (1977) Developing human: clinically oriented embryology. Saunders, Philadelphia
Oberthuer A, Hero B, Spitz R, Berthold F, Fischer M (2004) The tumor-associated antigen PRAME is universally expressed in high-stage neuroblastoma and associated with poor outcome. Clin Cancer Res 10:4307–4313. https://doi.org/10.1158/1078-0432.CCR-03-0813
Oehler VG, Guthrie KA, Cummings CL, Sabo K, Wood BL, Gooley T et al (2009) The preferentially expressed antigen in melanoma (PRAME) inhibits myeloid differentiation in normal hematopoietic and leukemic progenitor cells. Blood 114:3299–3308. https://doi.org/10.1182/blood-2008-07-170282
Paila U, Chapman BA, Kirchner R, Quinlan AR (2013) GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput Biol 9:e1003153. https://doi.org/10.1371/journal.pcbi.1003153
Priest JR, Watterson J, Strong L, Huff V, Woods WG, Byrd RL et al (1996) Pleuropulmonary blastoma: a marker for familial disease. J Pediatr 128:220–224
Pugh TJ, Yu W, Yang J, Field AL, Ambrogio L, Carter SL et al (2014) Exome sequencing of pleuropulmonary blastoma reveals frequent biallelic loss of TP53 and two hits in DICER1 resulting in retention of 5p-derived miRNA hairpin loop sequences. Oncogene 33:5295–5302. https://doi.org/10.1038/onc.2014.150
Rakheja D, Chen KS, Liu Y, Shukla AA, Schmid V, Chang TC et al (2014) Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat Commun 2:4802. https://doi.org/10.1038/ncomms5802
Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, Korbel JO (2012) DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28:i333–i339. https://doi.org/10.1093/bioinformatics/bts378
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. https://doi.org/10.1093/nar/gkv007
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Russell JP, Lodge EJ, Andoniadou CL (2018) Basic research advances on pituitary stem cell function and regulation. Neuroendocrinology 107:196–203. https://doi.org/10.1159/000488393
Sahakitrungruang T, Srichomthong C, Pornkunwilai S, Amornfa J, Shuangshoti S, Kulawonganunchai S et al (2014) Germline and somatic DICER1 mutations in a pituitary blastoma causing infantile-onset Cushing’s disease. J Clin Endocrinol Metab 99:E1487-1492. https://doi.org/10.1210/jc.2014-1016
Scheithauer BW, Horvath E, Abel TW, Robital Y, Park SH, Osamura RY et al (2012) Pituitary blastoma: a unique embryonal tumor. Pituitary 15:365–373. https://doi.org/10.1007/s11102-011-0328-x
Scheithauer BW, Kovacs K, Horvath E, Kim DS, Osamura RY, Ketterling RP et al (2008) Pituitary blastoma. Acta Neuropathol 116:657–666. https://doi.org/10.1007/s00401-008-0388-9
Seki M, Yoshida K, Shiraishi Y, Shimamura T, Sato Y, Nishimura R et al (2014) Biallelic DICER1 mutations in sporadic pleuropulmonary blastoma. Cancer Res 74:2742–2749. https://doi.org/10.1158/0008-5472.CAN-13-2470
Srirangam Nadhamuni V, Korbonits M (2020) Novel insights into pituitary tumorigenesis: genetic and epigenetic mechanisms. Endocr Rev 41:821–846. https://doi.org/10.1210/endrev/bnaa006
Tetreault M, Bareke E, Nadaf J, Alirezaie N, Majewski J (2015) Whole-exome sequencing as a diagnostic tool: current challenges and future opportunities. Expert Rev Mol Diagn 15:749–760. https://doi.org/10.1586/14737159.2015.1039516
Thorvaldsdottir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. https://doi.org/10.1093/bib/bbs017
Wang Y, Chen J, Yang W, Mo F, Senz J, Yap D et al (2015) The oncogenic roles of DICER1 RNase IIIb domain mutations in ovarian Sertoli-Leydig cell tumors. Neoplasia 17:650–660. https://doi.org/10.1016/j.neo.2015.08.003
Zhu X, Gleiberman AS, Rosenfeld MG (2007) Molecular physiology of pituitary development: signaling and transcriptional networks. Physiol Rev 87:933–963. https://doi.org/10.1152/physrev.00006.2006
Funding
This project was supported by the Canadian Cancer Society Research Institute Innovation grant #702071 and the Canadian Institutes for Health Research grant FDN-148390 awarded to WDF and was partially supported by C17 funded by the Childhood Cancer Canada Foundation. LdK was a recipient of a Vanier Canada Graduate Scholarship and Delta Kappa Gamma World Fellowship. We thank Alan Spatz MD PhD, Andreas Papadakis PhD and the Georges & Olga Minarik Research Pathology Facility for technical support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nadaf, J., de Kock, L., Chong, AS. et al. Molecular characterization of DICER1-mutated pituitary blastoma. Acta Neuropathol 141, 929–944 (2021). https://doi.org/10.1007/s00401-021-02283-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-021-02283-6