Abstract
An expanded hexanucleotide repeat in the C9orf72 gene is the most common genetic cause of frontotemporal dementia and amyotrophic lateral sclerosis (c9FTD/ALS). We now report the first description of a homozygous patient and compare it to a series of heterozygous cases. The patient developed early-onset frontotemporal dementia without additional features. Neuropathological analysis showed c9FTD/ALS characteristics, with abundant p62-positive inclusions in the frontal and temporal cortices, hippocampus and cerebellum, as well as less abundant TDP-43-positive inclusions. Overall, the clinical and pathological features were severe, but did not fall outside the usual disease spectrum. Quantification of C9orf72 transcript levels in post-mortem brain demonstrated expression of all known C9orf72 transcript variants, but at a reduced level. The pathogenic mechanisms by which the hexanucleotide repeat expansion causes disease are unclear and both gain- and loss-of-function mechanisms may play a role. Our data support a gain-of-function mechanism as pure homozygous loss of function would be expected to lead to a more severe, or completely different clinical phenotype to the one described here, which falls within the usual range. Our findings have implications for genetic counselling, highlighting the need to use genetic tests that distinguish C9orf72 homozygosity.
Similar content being viewed by others
Introduction
Frontotemporal dementia (FTD) is the most common form of early-onset dementia, second only to Alzheimer’s disease, and is characterised clinically by changes in personality or language impairment and pathologically by the progressive degeneration of the frontal and anterior temporal lobes [21, 24]. FTD shares numerous similarities at the genetic and neuropathological level with amyotrophic lateral sclerosis (ALS), a devastating neurodegenerative disorder in which the loss of motor neurons in brain and spinal cord causes progressive weakness and paralysis, ultimately leading to death from respiratory failure [12]. ALS and FTD can co-occur, and they have been proposed to be part of the same spectrum of disease [17].
An expanded hexanucleotide repeat in the C9orf72 gene has been identified recently as the most common known genetic cause of both FTD and ALS [9, 20, 25]. Whilst <33 hexanucleotide repeats occur in the healthy general population, with just 2 repeats being the most common form, C9orf72 ALS/FTD cases carry 800–4,400 repeats [5]. C9orf72 positive FTD (c9FTD) cases may show clinically typical FTD features and have been described to most commonly present with behavioural variant frontotemporal dementia, often with prominent psychiatric and amnestic symptoms [19].
Pathologically, c9FTD patients have unique characteristics, including p62-positive neuronal cytoplasmic inclusions (NCIs) in cerebellar and dentate fascia granule cells and pyramidal neurons of the hippocampus [19, 23, 26]. The pathogenic mechanisms by which the hexanucleotide repeat expansion causes disease are unclear and both gain- and loss-of-function mechanisms have been proposed to play a role [3, 9, 10, 22]. Here, we present a case of FTD with a homozygous C9orf72 hexanucleotide repeat expansion and compare with heterozygous cases. Clinical features, neuropathology and expression data that we describe below carry important implications for disease pathogenesis and genetic counselling.
Materials and methods
DNA extraction and genotyping
Genomic DNA was extracted from peripheral blood using the Nucleon BACC2 DNA extraction kit (RPN8502) following the supplied protocol. DNA concentrations were determined using a Nanodrop ND-1000 spectrophotometer, and adjusted to a working concentration of 20 ng/μl TE buffer.
Rs3849942 genotyping: The surrogate marker rs3849942, defining the haplotypes at risk of expansion, was genotyped by allelic discrimination using the 5′ nuclease assay in conjunction with Minor Groove Binding (MGB) probes. The custom-designed assay was performed on the SDS7500 Fast Real Time PCR system (ABI) and genotyping calls were made using software v2.0.6.
Hexanucleotide repeat number assessment
Hexanucleotide repeat number was assessed by repeat primed PCR and carried out as previously described [19]. Fragment length analysis was undertaken on an ABI 3730xl automated sequencer. Analysis of repeat primed PCR (rpPCR) electrophoretograms was performed using Peak Scanner v1.0 (ABI). In addition, repeat number was assessed by fluorescent labelled PCR followed by fragment length analysis on an Applied Biosystems (ABI) 3730xl automated sequencer as previously described [19].
Microsatellite analysis
Microsatellite analysis was performed using ten markers spanning approximately 13.1 Mb of genomic DNA centred on the C9orf72 gene. PCR amplicons were generated using fluorescently end-labelled primers for microsatellite markers D9S1814(VIC), D9S976(FAM), D9S171(NED), D9S1121(VIC), D9S169(FAM), D9S263(HEX), D9S270(FAM), D9S104(FAM), D9S147E(NED) and D9S761(FAM). DNA products were electrophoresed on an ABI 3730xl automated sequencer. Data were analysed using ABI GeneMapper software v4.0 [Applied Biosystems (ABI)].
Southern blotting
Adaptation of standard blotting methods included the probing of AluI/DdeI digested genomic DNA with an oligonucleotide hybridisation probe from Eurofins MWG Operon (Germany) that comprised five hexanucleotide repeats (GGGGCC)5 labelled 3′ and 5′ with digoxigenin (DIG). Further methods followed the DIG Application Manual [Roche Applied Science (RAS)], except for the supplementation of DIG Easy Hyb buffer with 100 μg/ml denatured fragmented salmon sperm DNA. Detection was carried out as recommended in the DIG Application Manual using CSPD ready-to-use (RAS) as chemiluminescent substrate visualised on fluorescent detection film (RAS). Hexanucleotide repeat number was estimated by visual interpolation using a plot of log10 base pair number against migration distance which was created in Excel (Microsoft) and subtraction of the wild-type allele fragment size (199 bp). Full methods have been previously reported [5].
For single probe Southern hybridisation, 5 μg genomic DNA were digested with EcoRI. The Roche DIG labelling and detection system was used with a 1 kb C9orf72 genomic DNA specific probe.
Pathological analysis
Analysis was undertaken according to the Queen Square Brain Bank protocol [16]. p62 (1:200, BD Transduction Laboratories), TDP-43 (amino acids 1-261, Abnova, 1:800), C9RANT [3] and ubiquilin-2 [6] were analysed. Using a semi-quantitative approach, the frequency of p62-positive neuronal cytoplasmic inclusions was compared with TDP-43-positive inclusions in the reported regions of the frontal cortex, temporal cortex, hippocampus and cerebellum.
Semi-quantitative analysis of p62 and TDP-43-positive lesions was performed using a five-tiered semi-quantitative grading scale. The scoring of the pathological lesions was as follows: ‘0’ for the absence of p62-positive neuronal cytoplasmic inclusions (NCIs) and neuronal intranuclear inclusions (NIIs); ‘+’ for 1–5 inclusions present in an average of at least five microscopic fields using a 20× objective; ‘++’ for 6–10; ‘+++’ for between 11 and 20 inclusions; ‘++++’ for >20 lesions.
RNA extraction and transcription analysis
RNA extraction was performed using the miRNeasy Kit (Qiagen) and RNA quality was evaluated using the Agilent 2100 Bioanalyzer. Reverse transcription was performed using the QuantiTect kit (Qiagen) and real-time quantitative RT-PCR (qPCR) was performed using Taqman probes according to manufacturer’s instructions (Applied Biosytems Primers and probes were designed in order to specifically detect the V1, V2 and V3 C9orf72 isoforms (Supplementary table 1). Dilution curve experiments using the V1 and V3 C9orf72 primers vs beta-actin showed comparable efficiencies. Reactions were performed in triplicate and gene expression values were normalised using the housekeeping genes ACTB and GAPDH. Relative quantification of gene expression was calculated via the comparative threshold cycle (ddCt) method. Regression analysis was performed by plotting the expression values vs the number of mutated alleles using Graphpad Prism v5.03.
Results
Patient report
A 45-year-old man originally from Turkey presented to a cognitive disorders clinic with a 2-year history of change in behaviour and cognitive impairment. He had become apathetic with increasing irritability and his behaviour was described as having become increasingly childish. Over the following 2 years prior to presentation he rapidly deteriorated such that when assessed he was severely apathetic and speech output was minimal. He was unable to follow more than simple commands and did not recognise his family. He required assistance with dressing and eating and was doubly incontinent.
The proband’s mother and father were first cousins, their fathers being brothers. Both parents developed an early-onset dementia, and a number of other family members developed dementia, with an autosomal dominant compatible transmission (Fig. 1a).
Neurological examination revealed an MMSE of 9 and relatively global cognitive impairment with formal neuropsychometric testing. The proband was echolalic. During testing he was perseverative and disinhibited. He had a mild extrapyramidal syndrome with bilateral limb bradykinesia and rigidity with stimulus-sensitive myoclonus and limb apraxia. He had symmetrically brisk reflexes with downgoing plantar responses. Limb power and co-ordination were normal with no wasting or fasciculations—EMG was not performed. He continued to progress rapidly with increasingly disinhibited and aggressive behaviour and died less than a year later.
Cerebro-spinal fluid (CSF) examination revealed tau levels of 171 pg/ml (0–320) and Aβ1-42 of 180 pg/ml (220–2,000). Oligoclonal bands were negative and CSF was acellular. EEG was unremarkable. A brain MRI showed generalised atrophy with frontal and medial temporal predominance without any clear asymmetry (Fig. 1b).
Genetic analysis
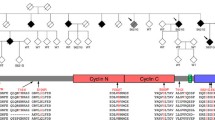
The patient’s DNA was sequenced to screen the PRNP, APP, PSEN1, PSEN2, GRN, and MAPT, genes, all of which were negative for mutations, and for C9orf72 hexanucleotide repeat number by repeat primed PCR, which showed a minimum of 40 repeats and a positive expansion pattern (Fig. 2a). The presence of a large expansion was confirmed by Southern blotting on DNA from peripheral lymphocytes using a GGGGCC5 oligonucleotide probe that targets the expansion [5], revealing a maximum of 3,067 repeats, minimum 830 (mode of 2,297 repeats, Fig. 2c). Assessment of repeat number of the presumed non-expanded allele by fluorescent PCR appeared to fail repeatedly, not revealing any bands despite test sensitivity proven from 2 to 32 repeats [5]. Results of haplotype analysis using 10 highly polymorphic microsatellite markers spanning 13.1 Mb centred on C9orf72 showed homozygosity at all markers and genotyping of rs3849942 demonstrated homozygosity for the adenosine nucleotide at this polymorphic site, typically linked with expanded alleles (Fig. 2b). These results pointed to the presence of a homozygous C9orf72 expansion and indeed, Southern blotting performed using a single-copy probe detecting the sequence adjacent to the repeat showed lack of a normal allele, confirming homozygosity of the C9orf72 expansion (Fig. 2d).
Presence of a homozygous hexanucleotide expansion in C9orf72. a Results of repeat primed PCR for C9orf72 expansion demonstrating the saw-tooth pattern, typical of the pathological expansion. Repeats are measurable up to 40 hexanucleotide repeats. The size of fluorescently labelled DNA amplicons is shown in base pairs (bp) against the expected asymptotic decay in fluorescence measured in arbitrary units. The GS500 size standard can also be seen with red peaks at 300, 340, 350, 400, 450, 490 and 500 bp. b Results of genetic analysis of the proband that demonstrate homozygosity at all microsatellite marker positions and at rs3849942. Positions of markers are given in megabases relative to chromosome and C9orf72 position. c Southern blot analysis on blood-derived DNA using a probe directed to the hexanucleotide repeat (GGGGCC)5 shows the presence of an expansion in the proband (C9 hom). The expansion is calculated to have a maximum of 3,067, a minimum 830 and mode of 2,297 repeats and is indistinguishable from a C9orf72 heterozygous case (C9 het). d Southern blot analysis on blood and brain (asterisk) derived DNA using a single-copy probe detecting the sequence adjacent to the repeat. The normal allele runs as an 8-kb band and is detectable in controls and C9orf72 expansion heterozygous cases, but is absent in the proband (C9 hom)
Neuropathology analysis of C9orf72 homozygous and heterozygous cases
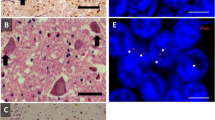
Neuropathological examination revealed a reduced brain weight (1,015 g), with frontoparietal atrophy. The corpus callosum, caudate, globus pallidus, thalamus, amygdala and hippocampus were all reduced in bulk. There was also pallor of the substantia nigra and locus coeruleus. However, the cerebellar white matter and dentate nucleus appeared normal. Microscopic assessment identified morphological features consistent with FTLD-TDP type A including TDP-43-positive neuronal cytoplasmic inclusions (NCIs), neuronal intranuclear inclusions (NIIs) and short neuritic threads. TDP-43-positive neurites and oligodendroglial inclusions were found in the deep white matter of the frontal cortex (Supplementary figure 1). No spinal cord was available for study; however, motor neurons of the 7th and 12th nerve nuclei were available for pathological assessment and showed no TDP-43 or p62-positive inclusions. There was no evidence of hippocampal sclerosis, whilst AT8 immunohistochemistry showed neuropil threads and neurofibrillary tangles in the transentorhinal and entorhinal cortices in keeping with Braak and Braak stage II. p62-positive NCIs were evident not only in the hippocampus and cerebellum, a consistent feature of C9orf72 expansion cases, but were also present in the majority of neurons in the middle and deeper cortical layers (Table 1; Fig. 3b, f). In keeping with previous descriptions of C9orf72 expansion cases ubiquilin-2 inclusions were found in both cerebellum and hippocampus (Fig. 3c, g). Further, we investigated the presence of dipeptides derived from the unconventional translation of the hexanucleotide repeat and found these to be present in hippocampal and cerebellar neurons (Fig. 3d, h). In all regions the number of p62-positive NCIs outnumbered the TDP-43 pathology. The p62 and TDP43 pathology was compared to five C9orf72 heterozygous cases and assessed as severe but within the range of C9orf72 cases (Table 1; Fig. 4a).
Immunohistochemical analysis of the neuronal cytoplasmic inclusions in the C9orf72 homozygous case. The ‘star-like’ neuronal cytoplasmic inclusions observed in the granule cell layer of the hippocampus (a–d) and the cerebellum (e–h) have been shown to be positive to varying degrees with several antibodies to proteins shown to be associated with C9orf72 cases. TDP-43 immunohistochemistry showed compact neuronal cytoplasmic inclusions (a arrows) together with the normal neuronal nuclear staining pattern. Only the normal nuclear staining pattern was observed in the cerebellum with the TDP-43 antibody (e). p62 immunohistochemisty (b and f) demonstrated large numbers of ‘star-like’ inclusions in both the GCL (b) and cerebellum (f). Ubiquilin-2 has been shown to be present in the small ‘star-like’ inclusions in heterozygous C9orf72 cases. The homozygous presented here also demonstrated a similar staining pattern with inclusions found in the GCL (c) and cerebellum (g). The newly identified C9RANT protein was also shown to be present in a small proportion of inclusions in the GCL (d) and cerebellum (h arrows). Bar in a represents 20 μm in all panels
Histopathological features and C9orf72 expression. p62 immunohistochemistry in the granule cell layer (GCL) of the hippocampus (a–c) and cerebellum (d–f) demonstrating the number of inclusions observed in the heterozygous cases range from mild (a, d) to severe (b, e). The severity of the inclusions observed in the homozygous case (c, f) is shown in both the GCL and cerebellum. Bar represents 100 μm in all panels. Real-time quantitative RT-PCR expression analysis of the three known C9orf72 isoforms—V1, V2, V3 (g, h)—schematically represented in i. Hexanucleotide expansion (red triangle), exons (green), intron (black line). Linear regression analysis (g) between expression and number of normal alleles shows significance for V1 and V2. Reduction of expression in C9orf72 heterozygous cases is significant only for V1 (p = 0.03)
C9orf72 transcription analysis of C9orf72 isoforms
For quantitative RT-PCR analysis for the three known C9orf72 transcripts [9], we extracted RNA from the frontal cortex of the proband, three heterozygous C9orf72 FTD cases and four pathologically confirmed controls without neurodegenerative disease. Results, normalised to β-actin, showed a significant reduction to 57 % of transcript variant 1 (V1) in heterozygous cases versus controls, whereas the homozygous case was reduced further to 20 % of control expression. Transcript variants 2 and 3 (V2 and V3) also showed a reduction in heterozygous cases, although not statistically significant, and a further reduction, respectively, to 34 and 45 % in the homozygous case versus controls (Fig. 3b, c). Results were confirmed using GAPDH as a second endogenous control.
Discussion
We report the first FTD patient carrying a homozygous C9orf72 hexanucleotide repeat expansion. Our finding that the clinicopathological features were severe, but within the range of reported heterozygous cases, suggests that the condition is truly dominant; therefore, we provide evidence in favour of a gain-of-function mechanism. Whilst the affected sister with an age of onset of 21 could also be potentially homozygous for the C9orf72 expansion, it is highly unlikely that the patient’s mother and also the father’s son from his second (non-consanguineous) marriage, both with an onset in their 20s, were homozygous. These family members were not available for assessment.
We report Southern blotting for detection of C9orf72 expansions using two different approaches with the probe directed towards either the hexanucleotide repeat itself (Fig. 2c) or the adjacent genomic region (Fig. 2d). The former has the advantage of being more sensitive and is able to detect a significant amount of somatic mosaicism, whilst the latter has the advantage of detecting also the normal allele and therefore its absence in the setting of homozygosity. A size shift, possibly due to somatic instability, is documented between blood and brain DNA samples, as previously reported [5].
Three major C9orf72 transcripts have been described: V1, in which the hexanucleotide expansion is in the promoter region, and V2 and V3, in which the expansion lies in the first intron and is therefore transcribed [9]. Previous analyses in C9orf72 heterozygous cases showed a reduction in V1 and V2 [9, 10]. Consistent with these findings, our results indicate a reduction of all three transcripts in heterozygous cases, with a further reduction in the homozygous case; but importantly, we unequivocally show that transcripts derived from expansion-containing alleles are present and clearly detectable in the homozygous case. One study [9] observed a greater reduction in the V1 isoform than V2 and V3. Although our results show that all variants are reduced, we nonetheless observe a greater reduction in V1 compared to V2 and V3 in the homozygous case.
The pathogenic mechanism of the C9orf72 repeat expansion is unknown and both loss of function (LOF) [10] and gain of function [22] have been proposed to play a role. Although the two mechanisms are not mutually exclusive, the clinicopathological data along with the expression profile of the case presented here are consistent with a predominant gain-of-function mechanism. A pure LOF mutation would be expected to cause a much more severe clinical presentation and pathology in the homozygous state, which was not observed in this case. Indeed heterozygous LOF mutations in other neurodegenerative diseases present with radically different phenotypes when homozygous, whereas those associated with a toxic gain of function tend to be present with the same clinical syndrome, although in many cases with an earlier onset. Examples include heterozygous LOF mutations in the progranulin gene (PGRN) causing FTD but homozygous mutations causing a lysosomal storage disorder [4, 29]; mutations in GBA causing Parkinson’s disease in heterozygosity and Gaucher’s disease in homozygosity [1]; and mutations in TREM2 causing Alzheimer’s disease and Nasu–Hakola disease [11, 14]; however, homozygous mutations in gain-of-function neurodegenerative diseases such as inherited prion disease (PRNP), ALS (SOD1) and Huntington disease (HTT), manifest with severe phenotypes, that fall within the range of disease [13, 15, 28, 30]. Further, a severe form of disease, but with no additional clinical features, such as presented here, was described for homozygous cases of myotonic dystrophy types 1 and 2 (DM1 and DM2) [2, 27]. Importantly, these disorders are similar to c9FTD in that they are also caused by very long expansions of non-coding repeats, and functional evidence suggests they occur via a gain-of-function mechanism [7]. The severe pathology here reported in the C9orf72 homozygous case could be compatible with a dosage dependence of the GOF mechanism. Gain and loss of function have been shown to coexist in other neurodegenerative disorders [8, 18, 31, 32]. Given the decreased levels of all C9orf72 transcripts, further work is needed to assess the possible role of loss of function in C9orf72 ALS/FTD, but our results argue against a pure loss-of-function mechanism.
Lastly, C9orf72 is estimated to have a frequency of 1:700 in the UK population [5], thus making the predicted frequency of homozygous cases to be approximately 1:2 × 106. Although we present a single case and clinical variability is likely to occur, our results show that homozygous C9orf72 expansion is viable and that it can present as typical FTD. Genetic diagnostic tests are already available for C9orf72 and are carried out by performing both repeat-primed PCR and Southern blotting. Our results underline the importance of performing Southern blots in the diagnostic test, in order to detect potential homozygous cases. Although homozygosity may or may not have prognostic implications, it carries huge implications for genetic counselling. In summary, we provide the first report of a homozygous C9orf72 repeat expansion FTD case, which provides new insight into disease pathogenesis and important implications for genetic testing.
References
Aharon-Peretz J, Rosenbaum H, Gershoni-Baruch R (2004) Mutations in the glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N Engl J Med 351(19):1972–1977
Akbas F, Serdaroglu P, Deymeer F, Aysal F, Erginel-Unaltuna N (2001) Molecular and clinical study of two myotonic dystrophy homozygotes. J Med Genet 38(11):e40
Ash PEA, Bieniek KF, Gendron TF et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. doi:10.1016/j.neuron.2013.02.004
Baker M, Mackenzie IR, Pickering-Brown SM et al (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442(7105):916–919
Beck J, Poulter M, Hensman D et al (2013) Large C9orf72 Hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am J Hum Genet. doi:10.1016/j.ajhg.2013.01.011
Brettschneider J, Van Deerlin VM, Robinson JL et al (2012) Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol 123(6):825–839
Cooper TA, Wan L, Dreyfuss G (2009) RNA and disease. Cell 136(4):777–793
Crespo-Barreto J, Fryer JD, Shaw CA, Orr HT, Zoghbi HY (2010) Partial loss of ataxin-1 function contributes to transcriptional dysregulation in spinocerebellar ataxia type 1 pathogenesis. PLoS Genet 6(7):e1001021
DeJesus-Hernandez M, Mackenzie IR, Boeve BF et al (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72(2):245–256
Gijselinck I, Van Langenhove T, van der Zee J et al (2012) A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol 11(1):54–65
Guerreiro R, Wojtas A, Bras J et al (2013) TREM2 variants in Alzheimer’s disease. N Engl J Med 368(2):117–127
Haverkamp LJ, Appel V, Appel SH (1995) Natural history of amyotrophic lateral sclerosis in a database population. Validation of a scoring system and a model for survival prediction. Brain 118(Pt 3):707–719
Hayward C, Brock DJ, Minns RA, Swingler RJ (1998) Homozygosity for Asn86Ser mutation in the CuZn-superoxide dismutase gene produces a severe clinical phenotype in a juvenile onset case of familial amyotrophic lateral sclerosis. J Med Genet 35(2):174
Jonsson T, Stefansson H, Steinberg S et al (2013) Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med 368(2):107–116
Kato M, Aoki M, Ohta M, Nagai M, Ishizaki F, Nakamura S, Itoyama Y (2001) Marked reduction of the Cu/Zn superoxide dismutase polypeptide in a case of familial amyotrophic lateral sclerosis with the homozygous mutation. Neurosci Lett 312(3):165–168
Lashley T, Rohrer JD, Bandopadhyay R et al (2011) A comparative clinical, pathological, biochemical and genetic study of fused in sarcoma proteinopathies. Brain 134(Pt 9):2548–2564
Lillo P, Hodges JR (2009) Frontotemporal dementia and motor neurone disease: overlapping clinic-pathological disorders. J Clin Neurosci 16(9):1131–1135
Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY (2008) Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature 452(7188):713–718
Mahoney CJ, Beck J, Rohrer JD et al (2012) Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: clinical, neuroanatomical and neuropathological features. Brain 135(Pt 3):736–750
Majounie E, Renton AE, Mok K et al (2012) Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol 11(4):323–330
McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ (2001) Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick’s Disease. Arch Neurol 58(11):1803–1809
Mori K, Weng S-M, Arzberger T et al (2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. doi:10.1126/science.1232927
Murray ME, DeJesus-Hernandez M, Rutherford NJ et al (2011) Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol 122(6):673–690
Neary D, Snowden JS, Gustafson L et al (1998) Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology 51(6):1546–1554
Renton AE, Majounie E, Waite A et al (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72(2):257–268
Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, Rogelj B, Al-Chalabi A, Hortobágyi T, Shaw CE (2011) p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol 122(6):691–702
Schoser BGH, Kress W, Walter MC, Halliger-Keller B, Lochmüller H, Ricker K (2004) Homozygosity for CCTG mutation in myotonic dystrophy type 2. Brain 127(Pt 8):1868–1877
Simon ES, Kahana E, Chapman J, Treves TA, Gabizon R, Rosenmann H, Zilber N, Korczyn AD (2000) Creutzfeldt–Jakob disease profile in patients homozygous for the PRNP E200K mutation. Ann Neurol 47(2):257–260
Smith KR, Damiano J, Franceschetti S et al (2012) Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet 90(6):1102–1107
Squitieri F, Gellera C, Cannella M et al (2003) Homozygosity for CAG mutation in Huntington disease is associated with a more severe clinical course. Brain 126(4):946–955
Winklhofer KF, Tatzelt J, Haass C (2008) The two faces of protein misfolding: gain- and loss-of-function in neurodegenerative diseases. EMBO J 27(2):336–349
Zuccato C, Valenza M, Cattaneo E (2010) Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev 90(3):905–981
Acknowledgments
This work was funded by the Medical Research Council (UK). We are grateful for the support of individuals and families involved. P.F. is funded by a Medical Research Council/Motor Neurone Disease Association Lady Edith Wolfson Fellowship. J.D.W. is supported by a Wellcome Trust Senior Clinical Fellowship (091673/Z/10/Z). T.L. is supported by an Alzheimer’s Research UK (ARUK) fellowship. A.M.I. was funded by ARUK and the MHMS General Charitable Trust. We thank the Queen Square Brain Bank for Neurological Disorders, UCL Institute of Neurology, London, for providing tissue. This work was undertaken at University College London Hospitals/University College London, which received a proportion of funding from the Department of Health’s National Institute for Health Research Biomedical Research Centres funding scheme.
Conflict of interest
No author had any conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
401_2013_1147_MOESM2_ESM.tif
Supplementary figure 1 TDP-43 immunohistochemistry in the deep white matter of the frontal cortex. TDP-43 positive neurites (a, arrow) and oligodendroglial inclusions (b, arrows) are observed in the deep white matter of the frontal cortex. Bar in a represents 20 μm (TIFF 300 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Fratta, P., Poulter, M., Lashley, T. et al. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol 126, 401–409 (2013). https://doi.org/10.1007/s00401-013-1147-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-013-1147-0