Abstract
In sporadic Alzheimer’s disease (AD), neurofibrillary lesion formation is preceded by extensive post-translational modification of the microtubule associated protein tau. To identify the modification signature associated with tau lesion formation at single amino acid resolution, immunopurified paired helical filaments were isolated from AD brain and subjected to nanoflow liquid chromatography–tandem mass spectrometry analysis. The resulting spectra identified monomethylation of lysine residues as a new tau modification. The methyl-lysine was distributed among seven residues located in the projection and microtubule binding repeat regions of tau protein, with one site, K254, being a substrate for a competing lysine modification, ubiquitylation. To characterize methyl lysine content in intact tissue, hippocampal sections prepared from post mortem late-stage AD cases were subjected to double-label confocal fluorescence microscopy using anti-tau and anti-methyl lysine antibodies. Anti-methyl lysine immunoreactivity colocalized with 78 ± 13% of neurofibrillary tangles in these specimens. Together these data provide the first evidence that tau in neurofibrillary lesions is post-translationally modified by lysine methylation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is defined in part by the appearance of intracellular inclusions composed of the microtubule associated protein tau [35]. The mechanisms that drive tau lesion formation in the highly prevalent sporadic form of AD are not fully understood, but appear to involve abnormal post-translational modifications (PTMs) that influence tau function, stability, and aggregation propensity [45]. For example, hyperphosphorylation of tau protein on certain hydroxy amino acids favors lesion formation by dissociating tau from its microtubule binding partner [4, 7] and by directly raising its rate and extent of aggregation [1, 9, 48]. Although tau phosphorylation state is mediated directly by phosphotransferases, it also is modulated by competing modifications on hydroxy amino acids such as O-linked β-N-acetylglucosaminylation (O-GlcNAcylation) [40]. The reciprocal relationship between these tau modifications is leveraged by O-GlcNAcase inhibitors, which by increasing O-GlcNAcylation, lower phosphorylation stoichiometry and depress neurofibrillary lesion formation [65]. In addition to hydroxy amino acids, Lys residues are modified on tau protein, and these too can influence tau metabolism and aggregation. For example, ubiquitylation of tau at Lys residues modulates intracellular tau levels [53, 61], the magnitude of which affects both nucleation and extension phases of the aggregation reaction [12]. Together these observations suggest that tau aggregation is under complex regulatory control that involves crosstalk among diverse and sometimes competing PTMs.
To gain insight into the tau PTM signature most closely associated with neurofibrillary lesion formation at single amino acid resolution, we have begun mapping modifications on authentic, paired helical filaments (PHFs) isolated from AD brain using mass spectrometry methods, with special emphasis on Lys modifications [13]. Preliminary analysis identified K254, K311, and K353 within the tau microtubule binding repeat region as ubiquitylation sites that were at least partially occupied in PHFs [13]. Recently, acetylation was discovered as another Lys-directed tau modification associated with tau-bearing lesions in AD and frontotemporal dementia [11, 46]. Tau acetylated in vitro resulted in occupation of diverse sites that overlapped with those we had determined to be ubiquitylated in disease [46]. Therefore, acetylation is another candidate modification for regulating tau turnover indirectly through the ubiquitin–proteasome system [46]. Moreover, in other biochemical pathways, such as histone-mediated control of gene expression, certain acetylated Lys residues can alternatively be methylated, contributing to complex cross-talk among Lys and hydroxyl amino acid modifications [36]. These observations suggest that the web of tau PTMs is potentially complex, with Lys-directed modifications playing key regulatory roles with respect to rates of tau turnover and aggregation.
Here we extend our characterization of PHF-tau using mass spectrometry methods by expanding search criteria to include both acetyl- and methyl-lysine modifications. Although we found no evidence for tau acetylation/trimethylation at the level of detection available in our datasets, the results show that Lys monomethylation is a widespread PHF-tau modification. These data reveal tau methylation as a new tau PTM accompanying PHF deposition in vivo.
Materials and methods
Subjects and tissue preparation
This study used only archival, de-identified post mortem brain tissue samples from autopsies performed with informed consent of each patient or relative via procedures approved by the relevant institutional committees (University of Rochester, USA). Paraformaldehyde-fixed brain tissue was obtained from six elderly subjects with a clinical diagnosis of AD [mean age 80 ± 10 years (SD)] that was confirmed on neuropathological evaluation in which the Consortium to Establish a Registry for AD (CERAD) age-adjusted criteria were met [47], and from four pathologically normal controls [mean age 62 ± 10 years (SD)] without history of neurologic or psychiatric disorders. All AD cases satisfied criteria for Braak stages V or VI, whereas none of the control cases met pathological criteria for AD [6].
Affinity purification of PHF-tau and enzymatic digestion
PHF-tau was isolated from pooled, late-stage (Braak stages V or VI) AD neocortical regions by immunoaffinity chromatography (MC1 monoclonal antibody) as described previously [30]. PHF-tau was digested in solution in the presence of 40% methanol with either trypsin (Promega) or Lys-C (Sigma) followed by phosphopeptide enrichment (Immobilized gallium, Thermo Fisher Scientific) as detailed earlier [13].
Reductive methylation of recombinant human 4R tau
Reductive methylation was done as described previously with minor modifications [18]. Briefly, lyophilized recombinant full-length human 2N4R tau (100 μg) was re-suspended in 100 μl of 0.1 M citrate buffer (pH 6) and methylated (room temperature for 2 h) in the presence of 0.1 M sodium cyanoborohydride and 20 mM formaldehyde. Reaction products were separated from reactants by spin dialysis in 100 mM ammonium bicarbonate, pH 7.8.
Liquid chromatography–tandem mass spectrometry (LC–MS/MS)
Mass spectrometric analysis of PHF-tau was performed using an LTQ ion trap mass spectrometer controlled by Xcalibur v.1.4 software (Thermo Electron) coupled online to a nanoflow XTreme Simple LC system (CVC Micro-Tech) as previously described [13]. Briefly, peptides were either loaded onto a trap column (Agilent Zorbax C18 guard column, or Michrom Bioresources peptide cap trap) or loaded directly into the sample loop with 95% solvent A (2% acetonitrile, 0.1% formic acid) and 5% solvent B (95% acetonitrile, 0.1% formic acid). A 60 min linear gradient of 5–25% solvent B was used to elute the peptides from the reverse phase column (150 mm × 75 μm, 5 μm 300 Å C18; CVC Micro-Tech).
The mass spectrometer was equipped with a nanospray ionization source (Thermo Electron) using an uncoated 10 μm i.d. SilicaTip PicoTip nanospray emitter (New Objective). The spray voltage of the mass spectrometer was 2.0 kV and the heated capillary temperature was 200°C. The top five ions in each MS1 scan were selected for MS/MS fragmentation. After ions were selected for MS/MS fragmentation twice within 30 s, they were dynamically excluded for 30 s. An MS3 scan was triggered if, among the three most abundant ions in the MS/MS scan, a neutral loss of 98, 49, or 32.7 Da (corresponding to a loss of H3PO4 from 1+, 2+, or 3+ precursor ions, respectively) was detected. Other mass spectrometric data generation parameters were as follows: collision energy 24% (35% for MS3 scans), MS scan range 400–1,800 m/z, minimum MS signal intensity 500 counts, minimum MS/MS signal intensity 100 counts, and MS/MS activation time 120 ms (30 ms for MS3 scans).
Analysis of mass spectrometric data
Spectra were searched against a UniProtKB human protein database (version Oct 5, 2010; 20,259 reviewed sequences; 75,498 non-reviewed sequences) using Bioworks 3.3.1 SP1 with the SEQUEST algorithm. Search parameters included 1.5 amu peptide mass tolerance, 1.0 amu fragment tolerance, static Cys +57 (carbamidomethylation) modification and the following differential modifications: Met +16 (oxidation); Ser, Thr, Tyr +80 (phosphorylation); Ser, Thr −18 (dehydroalanine and 2-amino-dehydrobutyric acid, respectively); Lys, Asp, Glu +14 (monomethylation); Lys +28 (dimethylation); Lys +42 (trimethylation/acetylation); and Lys +114 (ubiquitylation). Fully enzymatic (trypsin or Lys-C) peptides with up to two missed cleavages and charge-state dependent cross correlation (XCorr) scores ≥1.5, 2.5, and 3.0 for 1+, 2+, and 3+ peptides, respectively, and ΔCn >0.1 were considered as initial positive identifications. All MS/MS and MS3 spectra of identified post-translationally modified peptides from the initial screening were subjected to manual verification.
All raw data from this study are freely available to the research community on our laboratory’s website, http://www.proteomeumb.org, which also serves as a data-sharing portal. The original raw data are also available from http://proteomecommons.org with the following Hash IDs: Tau trypsin digestion dataset: (Y297YWVSnnKcaY3jSnQkuCWj7tA1mls59uBBjjBpyqF5hOQ4lSmAuWNhvtdC1EQCsj5A7 XFM0/3zj9YvVUIJlIuuwcoAAAAAAAABrA==); Tau Lys-C digestion dataset: (pNmgHCue Fwyw3vtAgIdKGatW5G8weUl7ArA/Fg+OlChNsahHADGSEQp7iUAOzouc81DMfWpeq7Ii1pDcjOkrs1h9BHAAAAAAAAABpg==).
Antibodies
Anti-tau mouse monoclonal antibodies Tau5 [43] and AT8 [22] were obtained from Dr. L. I. Binder (Northwestern University Medical School) and Endogen (Woburn, MA), respectively. Rabbit polyclonal anti-methyl lysine (meK) antibody was obtained from Enzo Life Sciences (ADI-KAP-TF121; Plymouth Meeting, USA). Cy3-conjugated goat anti-rabbit IgG and Alexa Fluor 488-conjugated goat anti-mouse secondary antibodies were from Jackson Immuno Research Laboratories, Inc (West Grove, USA) and Invitrogen (Carlsbad, USA), respectively. Horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody used for Western Blot was from Kirkegaard and Perry Laboratories Inc (Gaithersburg, USA).
Immunohistochemistry
Coronal hippocampal tissue sections were cut (20 μm thickness) and processed for immunohistochemistry as described previously [17, 33]. Sections were rehydrated in PBST (2.7 mM KCl, 0.14 M NaCl, 8.1 mM Na2HPO4, 0.1% Tween-20, pH 7.4) and fixed (10 min) in ice cold methanol. After 3 × 5 min rinses in PBST, sections were blocked (1 h at 19°C) with 5% goat serum diluted in PBST, then incubated (overnight at 4°C) with primary antibodies diluted in 2.5% goat serum (Tau5, 1 μg/ml; Anti-meK, 0.3 μg/ml; AT8, 0.1 μg/ml). After washing in PBST (3 × 10 min), sections were incubated (1 h at 19°C) with fluorescent dye-labeled secondary antibodies (1.5 μg/ml Cy3-conjugated goat anti-rabbit IgG; 2 μg/ml Alexa Fluor 488-conjugated goat anti-mouse IgG). After washing in PBST (3 × 5 min), tissue was treated (10 min at 19°C) with 0.1% Sudan Black B (EM Diagnostics, Gibbstown, USA) in 80% ethanol to suppress lipofuscin autofluorescence [59]. Sections were then washed (2 × 5 min) in PBST and once for 5 min in 33 mM NaH2PO4,162 mM Na2HPO4, pH 7.4. Coverslips were mounted with Vectashield (Vector Laboratories, Burlingame, USA) and sealed with clear nail enamel. Labeled sections were viewed in a Leica TCS SL laser-scanning confocal system (40× oil HCX Plan Apo CS 0.75–1.25 NA or 100× oil HCX Plan Apo CS 0.70–1.40 NA objective lens) operated at wavelengths optimized for simultaneous detection of Alexa Fluor 488 (λex = 488 nm; λem = 500–530 nm), and Cy3 (λex = 543 nm; λem = 560–600 nm). Long wavelength fluorescence (λex = 633 nm; λem = 650–740 nm) also was monitored to assess autofluorescence intensity. Digital confocal images were captured at 1× and 3× digital zoom, and stored in Tagged Image Format. Both secondary antibodies displayed minimal non-specific staining under these conditions as determined by immunostaining in the absence of primary antibodies. For immunoadsorption assay, primary antibodies anti-meK (0.3 μg/ml) and AT8 (0.1 μg/ml) were diluted in 2.5% goat serum containing 2000-fold molar excess of recombinant 2N4R tau (0.52 mg/ml) and incubated overnight (4°C) with agitation. Pellets were separated by centrifugation at 30,000×g for 30 min, and the supernatant was used for immunohistochemistry as described above.
Analytical methods
The proportion of meK-positive lesions was estimated using the Wilson score method [17, 49]. Sufficient technical replicates (defined as Tau-positive bodies at least 7 μm in both length and width) were counted from at least five fields of each case so that the Wilson 95% confidence interval for colocalization was <15%. Overall mean colocalization was then calculated as the average of all six biological replicate means and reported ± standard deviation.
Results
PHF-tau is methylated in its N-terminal projection and microtubule binding domains
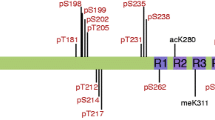
To identify sites of Lys modification in PHF-tau, previously collected MS datasets obtained from two independent preparations of authentic AD-brain derived PHFs (digested with either trypsin or Lys-C proteases; [13]) were interrogated using the SEQUEST database search algorithm programmed to identify unmodified Lys residues along with sites of monomethylation (K + 14), dimethylation (K + 28), trimethylation (K + 42), acetylation (also K + 42), and ubiquitylation (K + 114). Sequence coverage in these datasets included 25 out of the 44 Lys residues present in the longest form of human brain tau protein (2N4R tau, Fig. 1). This sequence coverage is based on only the unmodified, lysine-modified (methylated and ubiquitylated) and phosphorylated forms of PHF-tau. Although these search conditions confirmed three sites of ubiquitylation in the microtubule binding repeat region (K254, K311, and K353; [13]), no evidence for either K + 28 or K + 42 masses was found, indicating that dimethyl-, trimethyl-, and acetyl-lysine were not present in the coverage area at the level of detection available in our datasets. However, robust monomethylation was identified at seven sites distributed throughout the tau sequence (Table 1). Three of the sites (K163, K174, and K180) reside within the proline-rich region of the tau N-terminal projection domain, which mediates interactions with microtubule-associated proteins such as actin [27] and the Src homology three domain of plasma membrane-associated proteins including Src family kinases [37] and phospholipase Cγ [54]. In contrast, K254, K267, and K290 are part of the first and second repeats of the microtubule-binding domain. They reside within or adjacent to the core β-sheet region of filamentous tau in vivo [50] and flank the “PHF6*” sequence that modulates fibrillation rate of recombinant monomeric 4R tau in vitro [29, 39, 64]. Together these data reveal that PHF-tau is monomethylated in vivo, and that the major occupied sites distribute across protein segments known to mediate tau-protein interactions (including tubulin binding and self association).
Summary of modification sites identified by LC–MS/MS on immunopurified PHF-tau. The sequence shown is that of human 2N4R tau (NCBI accession number NP_005901). Bold sequence coverage; Red PHF6 and PHF6* motifs; dashed lines segments encoded by alternatively spliced exons 2, 3, and 10; underline repeat region (as defined in [23]); P Phosphorylated sites, U Ubiquitylated sites, me1 monomethylated sites identified from MS analysis reported herein and in [13]
To assess the relative abundances of methylated, ubiquitylated, and unmodified PHF-tau peptides, data were subjected to spectral counting, which measures the number of times a peptide is identified by MS/MS. Because spectral counts correlate linearly with protein abundance [41], they have been employed for relative quantification in many label-free proteomic studies [10, 51, 56, 57, 66]. Relative abundance was calculated by dividing the spectral count of a modified peptide by the sum of the spectral counts of all its forms (i.e., modified and unmodified). Results showed that the relative abundance of monomethylation varied among sites, from12% (K290) to as high as 67% (K180 and K267) (Fig. 2). The relative abundance of ubiquitylation also varied, from 1% (K254) to 33% (K311) (Fig. 2). These data indicate that Lys modification occupancies are substantial in PHF tau.
Relative abundance of PHF-tau methylation and ubiquitylation. Relative abundance was calculated based on the spectral counts of the modified peptide/(modified peptide + unmodified peptide). Only sites having total spectral counts >3 are shown. Inset quantifies K254 methylation (41%) and ubiquitylation (1%). K254 was the most abundant methylated site identified on PHF-tau (spectral count = 17) and the only site on PHF-tau identified in methylated and ubiquitylated forms
Lys methylation participates in competitive crosstalk with ubiquitylation
The sites of monomethylation identified above directly overlapped with sites previously identified as being acetylated in vitro (K174, K180; [46]) or ubiquitylated in vivo (K254; [13]), raising the issue of competitive crosstalk at these residues. Although no Lys acetylation was detected at these sites in our datasets, it was possible to quantify relative methylation and ubiquitylation of K254. Methylated K254 was identified in Lys-C peptide aa241-254 from manually verified MS/MS spectra (Fig. 3). Relative to the 1 + charge state y-ions of unmodified aa241–254 peptide (Fig. 3a), the y-ions of methylated aa241–254 were shifted by +14 Da (Fig. 3b), reflecting the mass of the methyl group (CH3) added to (and the H atom lost from) the ε-amino group on the K254 side chain. The appearance of the +14 Da shift was not likely an artifact of Fischer esterification (i.e., chemical methylation of free carboxylic acid groups in Asp and Glu; [32, 44]), because all MS/MS spectra were unambiguously assigned by SEQUEST (examples are shown in Supplemental Table 1). Moreover, care was taken to limit Fischer esterification conditions during sample preparation by maintaining the pH of digestion buffers >8, by avoiding exposure of samples to high concentrations of methanol in the presence of acetic acid, and by avoiding methanol as a co-solvent during LC separation of the peptides.
MS/MS characterization of K254 modification site. Spectra for peptide aa241–254 derived from Lys-C cleavage of PHF-tau identified in (a) unmodified and (b) methylated (asterisks site of methylation) forms. c Spectrum for tryptic peptide aa243–257 identified in ubiquitylated form (carets site of ubiquitylation). Trypsin did not cleave ubiquitylated aa243–257 at K254 owing to its conjugation to the C-terminal di-Gly fragment of ubiquitin
Ubiquitylated-K254 was identified in tryptic peptide aa243–257, which was resistant to protease cleavage at K254 owing to the isopeptide bond formed between ubiquitin and K254 (Fig. 3c). When aa241–254 and aa243–257 peptides were subjected to spectral counting, 41% of aa241–254 was monomethylated, whereas only1% of aa243–257 was ubiquitylated (Table 2; Fig. 2). These data suggest that monomethylation competes directly with ubiquitylation at K254, and that monomethylation of this site is far more abundant than ubiquitylation in PHF-tau (Fig. 2, inset).
Lys methylation and potential cross talk with phosphorylation
Although several of the monomethylation sites on PHF-tau lie within five amino acid residues of phosphorylation sites identified in the literature [25], only one peptide (aa258–267) was detected in our data sets that contained both a methylated lysine and a phosphorylated hydroxy amino acid. The phosphorylation site in this peptide, S262, functions as a regulatory gatekeeper for microtubule association, with site occupancy correlating with decreased binding affinity [38, 58]. Its proximity to K267 allowed us to detect relative site occupancy within a short peptide sequence. Unmodified aa258–267 was identified as a Lys-C derived peptide from its MS/MS spectrum (Fig. 4a). Relative to the 1+ charge state y-ions of this peptide, the y-ions of methylated aa258–267 peptide were shifted by +14 Da, reflecting the mass of the methyl group added to the ε-amino group of K267 (Fig. 4b). In contrast, the assigned y 7 - and y 8 -ions of aa258–267 phosphorylated at S262 were shifted by −18 Da, consistent with the dominant ion having undergone neutral loss of H3PO4 by β-elimination ([3]; Fig. 4c). Peptide aa258–267 also was detected in a doubly modified (phosphorylated and methylated) form. Consistent with the behavior of phosphopeptides, the MS/MS spectrum of this peptide was dominated by a precursor ion that had undergone β-elimination (Fig. 4d).
MS/MS identification of S262 phosphorylated and K267 methylated PHF-tau peptides. Spectra for PHF-tau derived peptide aa258–267 identified in a unmodified, b monomethylated (asterisks site of methylation), c Ser phosphorylated (at sign site of phosphorylation), and d doubly modified (asterisks site of methylation; at sign site of phosphorylation) forms
To quantify the modification signature of these peptides, their relative abundance was measured by spectral counting (Table 3). Results revealed that the singly modified methyl-K267 peptide was the most abundant form (spectral count = 6; 35% of total aa258–267 peptide spectral count), and relative abundance followed the pattern: methyl-K267 ~ methyl-K267/phospho-S262 > phospho-S262 ~ un-modified K267/S262. Thus monomethylation at K267 was a more abundant PHF-tau modification than was phosphorylation at S262, with the latter being found more frequently in the presence of K267 methylation than found alone.
Methyl-lysine immunoreactivity associates with the neurofibrillary lesions of AD
In AD, PHF-tau accumulates within neurofibrillary lesions associated with neuronal cell bodies (NFTs), neuronal processes (neuropil threads), and the dystrophic neurites of neuritic plaques [8]. To determine whether tau methylation correlated spatially with neurofibrillary pathology, sections of AD brain hippocampus were probed with a polyclonal antibody that binds meK-containing proteins along with well-characterized monoclonal anti-tau antibodies AT8 and Tau5 [5, 43] in double label format. These tau antibodies were chosen for analysis because their epitopes are present during all phases of NFT development, including the early pre- and intracellular-NFT stages [60]. The specificity of the anti-meK reagent was first tested on recombinant 2N4R tau preparations that were subjected to reductive methylation in vitro (see materials and methods). When subjected to SDS-PAGE, both non-methylated and methylated 2N4R tau migrated as single species, with the latter undergoing a band shift to 73 kDa (Fig. 5a). Rabbit polyclonal anti-meK antibody strongly labeled this species but not nonmethylated 2N4R tau (Fig. 5b). Binding specificity was further characterized by preadsorption assay, where antibodies were preincubated with 2000-fold molar excess of either non-methylated or methylated 2N4R tau before being used for immunohistochemical labeling of the hippocampal brain sections. When AT8 was preadsorbed under these conditions, neither non-methylated nor methylated tau preparations diminished its reactivity toward neurofibrillary lesions (detected by confocal immunofluorescence microcopy; Fig. 5c, f). These data were consistent with the established anti-phosphoepitope binding specificity of AT8 [22]. Anti-meK antibody also labeled neurofibrillary lesions in these sections, and like AT8, its labeling intensity was not affected by preadsorption with non-methylated 2N4R tau (Fig. 5d). However, labeling intensity was greatly diminished by pre-adsorption with methylated 2N4R tau (Fig. 5g). Together these data are consistent with the anti-meK antibody being selective for lysine methylated proteins.
Anti-meK antibody specificity. Aliquots of unmodified (−) and in vitro methylated (+) 2N4R tau were separated by SDS-PAGE (8% polyacrylamide) and either a stained with Coomassie Blue (500 ng tau proteins), or b subjected to immunoblot analysis with anti-meK antibody (100 ng tau protein). Molecular mass calibration markers (M) are shown in units of kDa. High-stoichiometry reductive methylation reduced recombinant 2N4R migration on SDS-PAGE. To further test specificity, antibodies AT8 (c, f; green channel) and anti-meK (d, g; red channel) were pre-adsorbed with either unmodified (c–e) or in vitro methylated (f–h) 2N4R tau, and then subjected to double-label confocal immunofluorescence microscopy on sections of AD hippocampus. Lesions labeled with both AT8 and anti-meK are marked by asterisks. AT8 labeling was unaffected by preadsorption with either unmodified or methylated tau, whereas anti-meK immunoreactivity was diminished by pre-adsorption with methylated (f–h; carets), but not unmodified tau (c–e)
Double-label confocal immunofluorescence studies were then extended to hippocampal brain sections prepared from four cognitively normal and six late-stage AD cases (case demographics are summarized in Table 4). In normal tissue, AT8, Tau5, and anti-meK antibody staining was diffuse and rarely colocalized (shown for AT8 and anti-meK antibody in Fig. 6a–c). In contrast, both AT8 (Fig. 6d–i) and Tau 5 (Fig. 6j–o) strongly labeled neurofibrillary lesions in AD hippocampal sections, including NFTs, neuropil threads, and neuritic plaques. At high magnification, NFTs immunolabeled with Tau5 displayed a fibrillar pattern throughout the lesions (Fig. 6m), whereas those labeled with AT8 also displayed a pattern of intense immunoreactivity enriched on the outer rims of the lesions (Fig. 6g). Rim staining, which has been seen previously with AT8 [28], was particularly conspicuous at low fluorescence gain. In contrast, anti-meK immunoreactivity appeared diffusely distributed throughout the sections (Fig. 6e, h, k, n), with the most intense labeling correlating with both AT8- and Tau5-stained lesions (Fig. 6d–o). In particular, the pattern of labeling with NFTs resembled that of Tau5, with fibrillar staining throughout the lesion (Fig. 6m–o). Although background fluorescence was generally too high to detect neuropil threads with the anti-meK antibody, colocalization with particularly large or intense dystrophic neurites was occasionally seen (Fig. 6j–l). NFT labeling was robust, however, and so colocalization of meK immunoreactivity with this lesion was quantified in hippocampus CA1 region. Results showed that the majority of NFTs labeled with anti-meK antibody in all six cases (Table 4). Overall, anti-meK colocalization with AT8-labeled NFTs averaged 78 ± 11% (SD, n = 6 cases) whereas colocalization with Tau5-labeled NFTs averaged 79 ± 16% (SD, n = 6 cases). Together these data show that the methylation of PHF-tau identified through mass spectrometry is widespread in this affected brain region, with most NFTs harboring anti-meK immunoreactivity in late-stage AD.
Anti-meK immunoreactivity colocalizes with neurofibrillary lesions in AD hippocampus. Double-label confocal images of hippocampal sections (CA1 region) stained with anti-tau mouse monoclonal antibodies AT8 (a–i; green channel) or Tau5 (j–o; green channel) along with the rabbit anti-meK antibody (red channel). In AD cases, low magnification images (40× objective,1× zoom) show colocalization of anti-meK immunoreactivity with NFTs (d–f; asterisks) and a neuritic plaque (j–l; arrows). High magnification images (100× objective, 3× zoom) show typical morphology of NFTs (g–i and m–o). Image overlays highlight pixel overlap between anti-meK and AT8 (d–i) and Tau5 (j–o) immunoreactivity. Anti-meK immunoreactivity colocalized extensively with NFTs in all fields examined. In contrast, AT8 and anti-meK immunoreactivity did not colocalize in hippocampal sections prepared from cognitively normal cases (a–c)
Discussion
Although certain familial tauopathies result from missense mutations in the tau gene (MAPT), AD pathogenesis is not associated with changes in tau amino acid sequence. Rather, tau lesion formation in sporadic AD is accompanied by PTMs that contribute to pathogenesis by modulating tau function, stability, and aggregation propensity. Because neurofibrillary lesion density correlates with neurodegeneration [24, 31] and cognitive decline [19, 20, 55], a high priority in the AD field is to identify the PTM signature that drives neurofibrillary lesion formation in sporadic disease. Here we found that PHF-tau isolated from neocortical areas is methylated at at least seven distinct sites, and that the modification is widespread among NFTs in the CA1 region of hippocampus in late stage AD. These findings suggest that tau methylation is a component of the PTM signature associated with PHF-tau.
The direct effects of Lys methylation on tau function are not known, but in other proteins this modification is reported to modulate protein–protein interaction by increasing the interaction radius of Lys side chains [34] and by increasing the entropic driving force for its burial away from solvent [62]. For these reasons, reductive methylation is used to improve the performance of protein crystallization in preparation for diffraction experiments [34]. The localization of methylated residues within the microtubule binding repeat region in proximity to the PHF6 and PHF6* nucleation centers makes Lys methylation a candidate modification for directly affecting normal tau interactions with binding partners such as microtubules as well as abnormal interactions such as aggregation and PHF formation.
In addition to direct effects, Lys methylation also may modulate protein function through cross-talk with other PTMs. In its simplest form, crosstalk involves direct competition for occupancy of any single amino acid residue side chain. Here we found that K254 from PHF-tau isolated from AD brains was either ubiquitylated or methylated, with the latter strongly predominating. These data indicate that K254 methylation must occur at the expense of ubiquitylation, and thus is a candidate modification for regulating protein degradation via the ubiquitin–proteasome system [46].
A second, more complex paradigm of crosstalk involves interactions between separate amino acid residues, where modification of one residue alters the ability of a second residue to bind its modifying enzyme. For example, tau phosphorylated by the microtubule affinity-regulating kinase 2 (PAR1/MARK2) is not recognized by the CHIP/Hsp90 E3 ligase complex and thus fails to be ubiquitylated or degraded [15]. Conversely, tau requires phosphorylation by GSK3β for efficient recognition and ubiquitylation by the CHIP/Hsp90 complex [61]. Here we found that K267 is methylated in PHF-tau, and that phosphorylation of S262 appears more frequently on peptides containing methylated rather than non-methylated K267. This observation raises the possibility of crosstalk between S262 phosphorylation, which strongly reduces the binding of tau to microtubules [4, 58], and K267 methylation. The impaired microtubule-binding phenotype associated with S262 phosphorylation may be important for AD pathogenesis since this site is occupied early in NFT formation [2]. Other sites of phosphorylation identified previously in PHF-tau [25] (but not found in our datasets) also lie within close proximity to methylation sites. These include T175, T181, S184, S185, S258, and S289. Moreover, one projection domain methylation site, K44, is adjacent to a predicted calpain-catalyzed cleavage site in tau [52]. These data suggest that tau methylation is positioned to potentially engage in cross talk with multiple post translational modifications. It will be important to establish the temporal relationship among these modifications and the aggregation of tau during neurofibrillary lesion formation in AD. Mass spectrometry can complement high throughput immunohistochemical detection methods, which are sensitive to modification context [16], in this effort.
Comparison with previous studies
Previous characterization of PHFs prepared by differential centrifugation using amino acid analysis reported a low unmodified Lys content relative to predicted tau composition, consistent with extensive Lys modification, but failed to detect meK [42]. The source of this discrepancy is not clear, but may relate to the sensitivity of amino acid analysis relative to MS and to the purity of the PHF preparations used for study.
Two recent studies reported that tau can be acetylated on Lys in vitro, resulting in increased aggregation propensity and inhibition of tau degradation [11, 46]. In vivo acetylation of K163, K174, K180 and K280 and accumulation within PHF-tau was claimed on the basis of antibody-based methods in tissue sections prepared from AD and frontotemporal lobar degeneration cases [11, 46]. On the basis of MS analysis, we did not detect tau acetylation even though residues K163, K174, K180 and K280 were resolved within our coverage area. These discrepancies could arise from any of several possibilities. One is that methylation occurs to a greater extent than acetylation. Consistent with this model, we found that K163, K174, and K180 were modified in PHF, but by monomethylation rather than acetylation. Combined with our detection of Lys ubiquitylation, these data suggest that the relative abundances of Lys methylation and ubiquitylation are likely higher than Lys acetylation in the PHF-tau preparation used herein. Another possibility is that PHF-tau contains substantial acetylation, but at sites distinct from those identified in vitro or that reside within our current coverage area. Additional analysis of PHF will be required to test this hypothesis. A third possibility is that immunopurified PHF represents a subfraction of tau aggregates with greater SDS-solubililty. For example, we find ([13]; Fig. 1) that immunopurified PHFs lack detectable phosphorylation at T175 and T181 that were reported qualitatively by others in some tissue/PHF preparations [2, 21, 26]. In addition, Y394 phosphorylation has been reported in PHF-tau [14, 63], but this modification is present at rather low relative abundance in the PHF-tau preparation used herein. It should be possible to resolve these possibilities in the future using mass spectrometry under conditions that provide quantitative information on modification occupancy.
In conclusion, we report biophysical evidence that seven Lys residues (K44, K163, K174, K180, K254, K267, and K290) in PHF-tau immunopurified from AD brain are monomethylated. The sites present opportunities for cross talk with other PTMs, including direct competition with ubiquitylation and acetylation, and indirect interaction with phosphorylation and proteolysis. It will be important to determine the functional and temporal relationships among these established modifications in modulating the accumulation of PHF-tau in AD and other age-related neurodegenerative diseases.
References
Alonso A, Zaidi T, Novak M, Grundke-Iqbal I, Iqbal K (2001) Hyperphosphorylation induces self-assembly of tau into tangles of paired helical filaments/straight filaments. Proc Natl Acad Sci USA 98:6923–6928
Augustinack JC, Schneider A, Mandelkow EM, Hyman BT (2002) Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer’s disease. Acta Neuropathol (Berl) 103:26–35
Beausoleil SA, Jedrychowski M, Schwartz D et al (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA 101:12130–12135
Biernat J, Gustke N, Drewes G, Mandelkow EM, Mandelkow E (1993) Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: distinction between PHF-like immunoreactivity and microtubule binding. Neuron 11:153–163
Braak H, Alafuzoff I, Arzberger T, Kretzschmar H, Del Tredici K (2006) Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol 112:389–404
Braak H, Braak E (1991) Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 82:239–259
Bramblett GT, Goedert M, Jakes R, Merrick SE, Trojanowski JQ, Lee VM (1993) Abnormal tau phosphorylation at Ser396 in Alzheimer’s disease recapitulates development and contributes to reduced microtubule binding. Neuron 10:1089–1099
Buee L, Bussiere T, Buee-Scherrer V, Delacourte A, Hof PR (2000) Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Brain Res Rev 33:95–130
Chang E, Kim S, Schafer KN, Kuret J (2011) Pseudophosphorylation of tau protein directly modulates its aggregation kinetics. Biochim Biophys Acta 1814:388–395
Choi H, Fermin D, Nesvizhskii AI (2008) Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics 7:2373–2385
Cohen TJ, Guo JL, Hurtado DE et al (2011) The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat Commun 2:252
Congdon EE, Kim S, Bonchak J, Songrug T, Matzavinos A, Kuret J (2008) Nucleation-dependent tau filament formation: the importance of dimerization and an estimation of elementary rate constants. J Biol Chem 283:13806–13816
Cripps D, Thomas SN, Jeng Y, Yang F, Davies P, Yang AJ (2006) Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J Biol Chem 281:10825–10838
Derkinderen P, Scales TM, Hanger DP et al (2005) Tyrosine 394 is phosphorylated in Alzheimer’s paired helical filament tau and in fetal tau with c-Abl as the candidate tyrosine kinase. J Neurosci 25:6584–6593
Dickey CA, Koren J, Zhang YJ et al (2008) Akt and CHIP coregulate tau degradation through coordinated interactions. Proc Natl Acad Sci USA 105:3622–3627
Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD (2011) Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol 21:53–58
Funk KE, Mrak RE, Kuret J (2011) Granulovacuolar degeneration (GVD) bodies of Alzheimer’s disease (AD) resemble late-stage autophagic organelles. Neuropathol Appl Neurobiol 37:295–306
Geoghegan KF (2001) Modification of amino groups. Curr Protoc Protein Sci Chapter 15, Unit 15.2
Ghoshal N, Garcia-Sierra F, Wuu J et al (2002) Tau conformational changes correspond to impairments of episodic memory in mild cognitive impairment and Alzheimer’s disease. Exp Neurol 177:475–493
Giannakopoulos P, Herrmann FR, Bussiere T et al (2003) Tangle and neuron numbers, but not amyloid load, predict cognitive status in Alzheimer’s disease. Neurology 60:1495–1500
Goedert M, Jakes R, Crowther RA et al (1994) Epitope mapping of monoclonal antibodies to the paired helical filaments of Alzheimer’s disease: identification of phosphorylation sites in tau protein. Biochem J 301(Pt 3):871–877
Goedert M, Jakes R, Vanmechelen E (1995) Monoclonal antibody AT8 recognises tau protein phosphorylated at both serine 202 and threonine 205. Neurosci Lett 189:167–169
Goedert M, Spillantini MG, Potier MC, Ulrich J, Crowther RA (1989) Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: differential expression of tau protein mRNAs in human brain. EMBO J 8:393–399
Gomez-Isla T, Price JL, McKeel DW Jr, Morris JC, Growdon JH, Hyman BT (1996) Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer’s disease. J Neurosci 16:4491–4500
Hanger DP, Anderton BH, Noble W (2009) Tau phosphorylation: the therapeutic challenge for neurodegenerative disease. Trends Mol Med 15:112–119
Hanger DP, Betts JC, Loviny TL, Blackstock WP, Anderton BH (1998) New phosphorylation sites identified in hyperphosphorylated tau (paired helical filament-tau) from Alzheimer’s disease brain using nanoelectrospray mass spectrometry. J Neurochem 71:2465–2476
He HJ, Wang XS, Pan R, Wang DL, Liu MN, He RQ (2009) The proline-rich domain of tau plays a role in interactions with actin. BMC Cell Biol 10:81
Ikonomovic MD, Abrahamson EE, Isanski BA et al (2006) X-34 labeling of abnormal protein aggregates during the progression of Alzheimer’s disease. Methods Enzymol 412:123–144
Iliev AI, Ganesan S, Bunt G, Wouters FS (2006) Removal of pattern-breaking sequences in microtubule binding repeats produces instantaneous tau aggregation and toxicity. J Biol Chem 281:37195–37204
Jicha GA, Bowser R, Kazam IG, Davies P (1997) Alz-50 and MC-1, a new monoclonal antibody raised to paired helical filaments, recognize conformational epitopes on recombinant tau. J Neurosci Res 48:128–132
Josephs KA, Whitwell JL, Ahmed Z et al (2008) Beta-amyloid burden is not associated with rates of brain atrophy. Ann Neurol 63:204–212
Jung SY, Li Y, Wang Y, Chen Y, Zhao Y, Qin J (2008) Complications in the assignment of 14 and 28 Da mass shift detected by mass spectrometry as in vivo methylation from endogenous proteins. Anal Chem 80:1721–1729
Kannanayakal TJ, Tao H, Vandre DD, Kuret J (2006) Casein kinase-1 isoforms differentially associate with neurofibrillary and granulovacuolar degeneration lesions. Acta Neuropathol 111:413–421
Kim Y, Quartey P, Li H et al (2008) Large-scale evaluation of protein reductive methylation for improving protein crystallization. Nat Methods 5:853–854
Kuret J (2007) Detection and reduction of neurofibrillary lesions. In: Smith HJ, Sewell RDE, Simons C (eds) Protein folding diseases: enzyme inhibitors and other agents as prospective therapies. CRC Press, Boca Raton, pp 287–324
Latham JA, Dent SY (2007) Cross-regulation of histone modifications. Nat Struct Mol Biol 14:1017–1024
Lee G, Newman ST, Gard DL, Band H, Panchamoorthy G (1998) Tau interacts with src-family non-receptor tyrosine kinases. J Cell Sci 111:3167–3177
Leger J, Kempf M, Lee G, Brandt R (1997) Conversion of serine to aspartate imitates phosphorylation-induced changes in the structure and function of microtubule-associated protein tau. J Biol Chem 272:8441–8446
Li W, Lee VM (2006) Characterization of two VQIXXK motifs for tau fibrillization in vitro. Biochemistry 45:15692–15701
Liu F, Iqbal K, Grundke-Iqbal I, Hart GW, Gong CX (2004) O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer’s disease. Proc Natl Acad Sci USA 101:10804–10809
Liu H, Sadygov RG, Yates JR 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76:4193–4201
Liu WK, Ksiezak-Reding H, Yen SH (1991) Abnormal tau proteins from Alzheimer’s disease brains. Purification and amino acid analysis. J Biol Chem 266:21723–21727
LoPresti P, Szuchet S, Papasozomenos SC, Zinkowski RP, Binder LI (1995) Functional implications for the microtubule-associated protein tau: localization in oligodendrocytes. Proc Natl Acad Sci USA 92:10369–10373
Lu A, Zougman A, Pudelko M et al (2009) Mapping of lysine monomethylation of linker histones in human breast and its cancer. J Proteome Res 8:4207–4215
Martin L, Latypova X, Terro F (2011) Post-translational modifications of tau protein: implications for Alzheimer’s disease. Neurochem Int 58:458–471
Min SW, Cho SH, Zhou Y et al (2010) Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron 67:953–966
Mirra SS, Heyman A, McKeel D et al (1991) The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD); Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 41:479–486
Necula M, Kuret J (2004) Pseudophosphorylation and glycation of tau protein enhance but do not trigger fibrillization in vitro. J Biol Chem 279:49694–49703
Newcombe RG (1998) Improved confidence intervals for the difference between binomial proportions based on paired data. Stat Med 17:2635–2650
Novak M, Kabat J, Wischik CM (1993) Molecular characterization of the minimal protease resistant tau unit of the Alzheimer’s disease paired helical filament. Embo J 12:365–370
Old WM, Meyer-Arendt K, Aveline-Wolf L et al (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics 4:1487–1502
Park SY, Ferreira A (2005) The generation of a 17 kDa neurotoxic fragment: an alternative mechanism by which tau mediates beta-amyloid-induced neurodegeneration. J Neurosci 25:5365–5375
Petrucelli L, Dickson D, Kehoe K et al (2004) CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet 13:703–714
Reynolds CH, Garwood CJ, Wray S et al (2008) Phosphorylation regulates tau interactions with Src homology 3 domains of phosphatidylinositol 3-kinase, phospholipase C gamma1, Grb2, and Src family kinases. J Biol Chem 283:18177–18186
Royall DR, Palmer R, Mulroy AR et al (2002) Pathological determinants of the transition to clinical dementia in Alzheimer’s disease. Exp Aging Res 28:143–162
Sardiu ME, Cai Y, Jin J et al (2008) Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci USA 105:1454–1459
Schmidt MW, Houseman A, Ivanov AR, Wolf DA (2007) Comparative proteomic and transcriptomic profiling of the fission yeast Schizosaccharomyces pombe. Mol Syst Biol 3:79
Schneider A, Biernat J, von Bergen M, Mandelkow E, Mandelkow EM (1999) Phosphorylation that detaches tau protein from microtubules (Ser262, Ser214) also protects it against aggregation into Alzheimer paired helical filaments. Biochemistry 38:3549–3558
Schnell SA, Staines WA, Wessendorf MW (1999) Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J Histochem Cytochem 47:719–730
Shimazaki M, Nakano H, Kobayashi K (2005) Correlation between tau phosphorylation sites and tangle morphology in Alzheimer’s disease. Psychogeriatrics 5:22–35
Shimura H, Schwartz D, Gygi SP, Kosik KS (2004) CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J Biol Chem 279:4869–4876
Sledz P, Zheng H, Murzyn K et al (2010) New surface contacts formed upon reductive lysine methylation: improving the probability of protein crystallization. Protein Sci 19:1395–1404
Tremblay MA, Acker CM, Davies P (2010) Tau phosphorylated at tyrosine 394 is found in Alzheimer’s disease tangles and can be a product of the Abl-related kinase, Arg. J Alzheimers Dis 19:721–733
von Bergen M, Friedhoff P, Biernat J, Heberle J, Mandelkow EM, Mandelkow E (2000) Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc Natl Acad Sci USA 97:5129–5134
Yuzwa SA, Vocadlo DJ (2009) O-GlcNAc modification and the tauopathies: insights from chemical biology. Curr Alzheimer Res 6:451–454
Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, Hettich RL, Samatova NF (2006) Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res 5:2909–2918
Acknowledgments
This work was supported by National Institutes of Health Grants MH59786 and AG25323 to A.Y. J.K is supported by AG14452. Y.W. and S.T. were supported by the Maryland Cigarette Restitution Fund. The access to the confocal microscope at Ohio State University was supported by Core Center grant P30-NS045758. The authors thank Diane Cripps for excellent technical assistance in the preparation of PHF-tau for LC–MS/MS analysis and also for the optimization of the LC–MS/MS platform used for the studies described herein. We thank Drs. Paul D. Coleman and Linda M. Callahan (University of Rochester, NY) for generous access to tissue resources.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding authors
Additional information
S.N. Thomas and K.E. Funk contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
401_2011_893_MOESM1_ESM.xls
SEQUEST .out file data for the MS/MS scans assigned to Tau aa25-44 and aa243-254 with unambiguous assignment of methylation to K44 and K254, respectively. The top-ranked peptides assigned to each spectrum are in bold, while the peptides with false-positive identifications (including putative methylated Asp and Glu residues) are in normal typeface (XLS 22 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Thomas, S.N., Funk, K.E., Wan, Y. et al. Dual modification of Alzheimer’s disease PHF-tau protein by lysine methylation and ubiquitylation: a mass spectrometry approach. Acta Neuropathol 123, 105–117 (2012). https://doi.org/10.1007/s00401-011-0893-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-011-0893-0