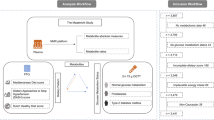
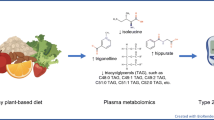
Abstract
Purpose
Dietary intakes are reflected in plasma by the presence of hundreds of exogenous metabolites and variations in endogenous metabolites. The exploration of diet-related plasma metabolic profiles could help to better understand the impact of overall diet on health. Our aim was to identify metabolomic signatures reflecting overall diet in women from the French general population.
Methods
This cross-sectional study included 160 women in the SU.VI.MAX cohort with detailed dietary data (≥ 10 24-h dietary records) selected according to their level of adherence to the French dietary recommendations, represented by the validated score mPNNS-GS; 80 women from the 10th decile of the score were matched with 80 women from the 1st decile. Plasma metabolomic profiles were acquired using untargeted UPLC-QToF mass spectrometry analysis. The associations between metabolomic profiles and the mPNNG-GS, its components and Principal Component Analyses-derived dietary patterns were investigated using multivariable conditional logistic regression models and partial correlations.
Results
Adherence to the dietary recommendations was positively associated with 3-indolepropionic acid and pipecolic acid (also positively associated with fruit and vegetable intake and a healthy diet)—2 metabolites linked to microbiota and inversely associated with lysophosphatidylcholine (LysoPC(17:1)), acylcarnitine C9:1 (also inversely associated with a healthy diet), acylcarnitine C11:1 and 2-deoxy-D-glucose. Increased plasma levels of piperine and Dihydro4mercapto-3(2H) furanone were observed in women who consumed a Western diet and a healthy diet, respectively. Ethyl-β-D-glucopyranoside was positively associated with alcohol intake. Plasma levels of LysoPC(17:1), cholic acid, phenylalanine-phenylalanine and phenylalanine and carnitine C9:1 decreased with the consumption of vegetable added fat, sweetened food, milk and dairy products and fruit and vegetable intakes, respectively.
Conclusion
This study highlighted several metabolites from both host and microbial metabolism reflecting the long-term impact of the overall diet.
Trial registration
SU.VI.MAX, clinicaltrials.gov NCT00272428. Registered 3 January 2006, https://clinicaltrials.gov/show/NCT00272428


Similar content being viewed by others
Abbreviations
- SU.VI.MAX:
-
SUpplémentation en Vitamines et Minéraux AntioXydants
- mPNNS-GS:
-
Modified Programme National Nutrition Santé- Guideline Score
- UPLC:
-
Ultrahigh-performance liquid chromatography
- LysoPC:
-
Lysophosphatidylcholine
- FFQ:
-
Food frequency questionnaire
- WHO:
-
World Health Organization
- NMR:
-
Nuclear magnetic resonance
- LC–MS:
-
Liquid chromatography mass spectrometry
- PCA:
-
Principal component analysis
- CVD:
-
Cardiovascular disease
- BMI:
-
Body mass index
- HCA:
-
Hierarchical clustering analysis
- OR:
-
Odds ratios
- SD:
-
Standard deviation
- CI:
-
Confidence interval
- FDR:
-
False discovery rate
- BH:
-
Benjamini-Hochberg
- PA:
-
Pipecolic acid
- IPA:
-
3-Indolepropionic acid
- 2DG:
-
2-Deoxy-D-glucose
- T2D:
-
Type 2 diabetes
- HEI:
-
Healthy eating index
- aMED:
-
Alternate mediterranean diet score
- HDI:
-
Healthy diet indicator
- BSD:
-
Baltic Sea diet
References
Jenab M, Slimani N, Bictash M et al (2009) Biomarkers in nutritional epidemiology: applications, needs and new horizons. HumGenet 125:507–525
Kaaks R, Ferrari P, Ciampi A et al (2002) Uses and limitations of statistical accounting for random error correlations, in the validation of dietary questionnaire assessments. Public Health Nutr 5:969–976
Sugar EA, Wang C-Y, Prentice RL (2007) Logistic regression with exposure biomarkers and flexible measurement error. Biometrics 63:143–151. https://doi.org/10.1111/j.1541-0420.2006.00632.x
Day N, McKeown N, Wong M et al (2001) Epidemiological assessment of diet: a comparison of a 7-day diary with a food frequency questionnaire using urinary markers of nitrogen, potassium and sodium. Int J Epidemiol 30:309–317. https://doi.org/10.1093/ije/30.2.309
Dragsted LO, Gao Q, Scalbert A et al (2018) Validation of biomarkers of food intake-critical assessment of candidate biomarkers. Genes Nutr 13:14. https://doi.org/10.1186/s12263-018-0603-9
Freedman LS, Tasevska N, Kipnis V et al (2010) Gains in statistical power from using a dietary biomarker in combination with self-reported intake to strengthen the analysis of a diet-disease association: an example from CAREDS. Am J Epidemiol 172:836–842. https://doi.org/10.1093/aje/kwq194
Jones DP, Park Y, Ziegler TR (2012) Nutritional metabolomics: progress in addressing complexity in diet and health. Annu Rev Nutr 32:183–202. https://doi.org/10.1146/annurev-nutr-072610-145159
González-Peña D, Brennan L (2019) Recent advances in the application of metabolomics for nutrition and health. Annu Rev Food Sci Technol 10:479–519. https://doi.org/10.1146/annurev-food-032818-121715
Playdon MC, Sampson JN, Cross AJ et al (2016) Comparing metabolite profiles of habitual diet in serum and urine. Am J Clin Nutr 104:776–789. https://doi.org/10.3945/ajcn.116.135301
Guertin KA, Moore SC, Sampson JN et al (2014) Metabolomics in nutritional epidemiology: identifying metabolites associated with diet and quantifying their potential to uncover diet-disease relations in populations. AmJClinNutr 100:208–217
Cespedes EM, Hu FB (2015) Dietary patterns: from nutritional epidemiologic analysis to national guidelines. Am J Clin Nutr 101:899–900. https://doi.org/10.3945/ajcn.115.110213
World Health Organization (WHO) (2017) Guideline: dietary patterns. WHO, Geneva, Switzerland
Brennan L (2017) Metabolomics: a tool to aid dietary assessment in nutrition. Curr Opin Food Sci 16:96–99. https://doi.org/10.1016/j.cofs.2017.09.003
Garcia-Perez I, Posma JM, Gibson R et al (2017) Objective assessment of dietary patterns by use of metabolic phenotyping: a randomised, controlled, crossover trial. Lancet Diabetes Endocrinol 5:184–195. https://doi.org/10.1016/S2213-8587(16)30419-3
Bondia-Pons I, Martinez JA, de la Iglesia R et al (2015) Effects of short- and long-term Mediterranean-based dietary treatment on plasma LC-QTOF/MS metabolic profiling of subjects with metabolic syndrome features: the metabolic syndrome reduction in Navarra (RESMENA) randomized controlled trial. Mol Nutr Food Res 59:711–728. https://doi.org/10.1002/mnfr.201400309
Andersen MB, Rinnan A, Manach C et al (2014) Untargeted metabolomics as a screening tool for estimating compliance to a dietary pattern. JProteomeRes 13:1405–1418
Acar E, Gürdeniz G, Khakimov B et al (2019) Biomarkers of individual foods, and separation of diets using untargeted LC-MS-based plasma metabolomics in a randomized controlled trial. Mol Nutr Food Res 63:1800215. https://doi.org/10.1002/mnfr.201800215
Vázquez-Fresno R, Llorach R, Urpi-Sarda M et al (2015) Metabolomic pattern analysis after mediterranean diet intervention in a nondiabetic population: a 1- and 3-year follow-up in the PREDIMED study. J Proteome Res 14:531–540. https://doi.org/10.1021/pr5007894
Hanhineva K, Lankinen MA, Pedret A et al (2015) Nontargeted metabolite profiling discriminates diet-specific biomarkers for consumption of whole grains, fatty fish, and bilberries in a randomized controlled trial. J Nutr 145:7–17. https://doi.org/10.3945/jn.114.196840
Lankinen M, Kolehmainen M, Jääskeläinen T et al (2014) Effects of whole grain, fish and bilberries on serum metabolic profile and lipid transfer protein activities: a randomized trial (Sysdimet). PLoS ONE 9:e90352. https://doi.org/10.1371/journal.pone.0090352
Stella C, Beckwith-Hall B, Cloarec O et al (2006) Susceptibility of human metabolic phenotypes to dietary modulation. J Proteome Res 5:2780–2788. https://doi.org/10.1021/pr060265y
Esko T, Hirschhorn JN, Feldman HA et al (2017) Metabolomic profiles as reliable biomarkers of dietary composition. Am J Clin Nutr 105:547–554. https://doi.org/10.3945/ajcn.116.144428
Peré-Trepat E, Ross AB, Martin F-P et al (2010) Chemometric strategies to assess metabonomic imprinting of food habits in epidemiological studies. Chemom Intell Lab Syst 104:95–100. https://doi.org/10.1016/j.chemolab.2010.06.001
Altmaier E, Kastenmüller G, Römisch-Margl W et al (2011) Questionnaire-based self-reported nutrition habits associate with serum metabolism as revealed by quantitative targeted metabolomics. Eur J Epidemiol 26:145–156. https://doi.org/10.1007/s10654-010-9524-7
Merz B, Frommherz L, Rist MJ et al (2018) Dietary pattern and plasma BCAA-variations in healthy men and women-results from the KarMeN study. Nutrients. https://doi.org/10.3390/nu10050623
Floegel A, von Ruesten A, Drogan D et al (2013) Variation of serum metabolites related to habitual diet: a targeted metabolomic approach in EPIC-Potsdam. Eur J Clin Nutr 67:1100–1108. https://doi.org/10.1038/ejcn.2013.147
Bouchard-Mercier A, Rudkowska I, Lemieux S et al (2013) The metabolic signature associated with the Western dietary pattern: a cross-sectional study. Nutr J 12:158. https://doi.org/10.1186/1475-2891-12-158
Almanza-Aguilera E, Urpi-Sarda M, Llorach R et al (2017) Microbial metabolites are associated with a high adherence to a Mediterranean dietary pattern using a 1H-NMR-based untargeted metabolomics approach. J Nutr Biochem 48:36–43. https://doi.org/10.1016/j.jnutbio.2017.06.001
Playdon MC, Moore SC, Derkach A et al (2017) Identifying biomarkers of dietary patterns by using metabolomics. Am J Clin Nutr 105:450–465. https://doi.org/10.3945/ajcn.116.144501
O’Sullivan A, Gibney MJ, Brennan L (2011) Dietary intake patterns are reflected in metabolomiprofilesc: potential role in dietary assessment studies. AmJClinNutr 93:314–321
Xu J, Yang S, Cai S et al (2010) Identification of biochemical changes in lactovegetarian urine using 1H NMR spectroscopy and pattern recognition. Anal Bioanal Chem 396:1451–1463. https://doi.org/10.1007/s00216-009-3338-z
Schmidt JA, Rinaldi S, Ferrari P et al (2015) Metabolic profiles of male meat eaters, fish eaters, vegetarians, and vegans from the EPIC-Oxford cohort. Am J Clin Nutr 102:1518–1526. https://doi.org/10.3945/ajcn.115.111989
Menni C, Zhai G, Macgregor A et al (2013) Targeted metabolomics profiles are strongly correlated with nutritional patterns in women. Metab Off J Metab Soc 9:506–514. https://doi.org/10.1007/s11306-012-0469-6
Bhupathiraju SN, Guasch-Ferré M, Gadgil MD et al (2018) Dietary patterns among Asian Indians living in the United States have distinct metabolomic profiles that are associated with cardiometabolic risk. J Nutr. https://doi.org/10.1093/jn/nxy074
Wei R, Ross AB, Su M et al (2018) Metabotypes related to meat and vegetable intake reflect microbial, lipid and amino acid metabolism in healthy people. Mol Nutr Food Res 62:1800583. https://doi.org/10.1002/mnfr.201800583
Gibbons H, Carr E, McNulty BA et al (2017) Metabolomic-based identification of clusters that reflect dietary patterns. Mol Nutr Food Res. https://doi.org/10.1002/mnfr.201601050
Sedlmeier A, Kluttig A, Giegling I et al (2018) The human metabolic profile reflects macro- and micronutrient intake distinctly according to fasting time. Sci Rep. https://doi.org/10.1038/s41598-018-30764-4
Guasch-Ferré M, Bhupathiraju SN, Hu FB (2018) Use of metabolomics in improving assessment of dietary intake. Clin Chem 64:82–98. https://doi.org/10.1373/clinchem.2017.272344
Hercberg S, Galan P, Preziosi P et al (2004) The SU.VI.MAX study: a randomized, placebo-controlled trial of the health effects of antioxidant vitamins and minerals. ArchInternMed 164:2335–2342
Hercberg S, Preziosi P, Briancon S et al (1998) A primary prevention trial using nutritional doses of antioxidant vitamins and minerals in cardiovascular diseases and cancers in a general population: the SU.VI.MAX study–design, methods, and participant characteristics. SUpplementation en VItamines et Mineraux AntioXydants. Control ClinTrials 19:336–351
Le Moullec N, Deheeger M, Preziosi P et al (1996) Validation du manuel photos utilisé pour l’enquête alimentaire de l’étude SU.VI.MAX (Validation of the food portion size booklet used in the SU.VI.MAX study). CahNutrDiet 31:158–164
Hercberg S (2005) Table de Composition SU.VI.MAX des Aliments. Les éditions INSERM/Economica, Paris
Lécuyer L, Dalle C, Lyan B et al (2019) Plasma metabolomic signatures associated with long-term breast cancer risk in the SU.VI.MAX prospective cohort. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. https://doi.org/10.1158/1055-9965.EPI-19-0154
Lécuyer L, Victor Bala A, Deschasaux M et al (2018) NMR metabolomic signatures reveal predictive plasma metabolites associated with long-term risk of developing breast cancer. Int J Epidemiol 47:484–494. https://doi.org/10.1093/ije/dyx271
Lécuyer L, Dalle C, Lefevre-Arbogast S, et al (2019) Diet-related metabolomic signature of long-term breast cancer risk using penalized regression: an exploratory study in the SU.VI.MAX cohort. Cancer Epidemiol Biomarkers Prev cebp.0900.2019. https://doi.org/10.1158/1055-9965.EPI-19-0900
Estaquio C, Kesse-Guyot E, Deschamps V et al (2009) Adherence to the French programme national nutrition Sante guideline score is associated with better nutrient intake and nutritional status. JAmDiet Assoc 109:1031–1041
Kesse-Guyot E, Touvier M, Henegar A et al (2011) Higher adherence to French dietary guidelines and chronic diseases in the prospective SU.VI.MAX cohort. EurJ ClinNutr 65:887–894
Lassale C, Galan P, Julia C et al (2013) Association between adherence to nutritional guidelines, the metabolic syndrome and adiposity markers in a French adult general population. PLoS ONE 8:e76349. https://doi.org/10.1371/journal.pone.0076349
Lassale C, Fezeu L, Andreeva VA et al (2005) (2012) Association between dietary scores and 13-year weight change and obesity risk in a French prospective cohort. Int J Obes 36:1455–1462. https://doi.org/10.1038/ijo.2011.264
Lavalette C, Adjibade M, Srour B et al (2018) Cancer-specific and general nutritional scores and cancer risk: results from the prospective NutriNet-Santé cohort. Cancer Res 78:4427–4435. https://doi.org/10.1158/0008-5472.CAN-18-0155
Pereira F, Martin JF, Joly C et al (2010) Development and validation of a UPLC/MS method for a nutritional metabolomic study of human plasma. Metabolomics 6:207–218
Giacomoni F, Le Corguille G, Monsoor M et al (2015) Workflow4Metabolomics: a collaborative research infrastructure for computational metabolomics. Bioinformatics 31:1493–1495. https://doi.org/10.1093/bioinformatics/btu813
Sumner LW, Amberg A, Barrett D et al (2007) Proposed minimum reporting standards for chemical analysis: chemical analysis working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3:211–221. https://doi.org/10.1007/s11306-007-0082-2
Cattell RB (1966) The scree test for the number of factors. Multivar BehavRes 1:245–276
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300
Fiedler K, Kutzner F, Krueger JI (2012) The long way from α-error control to validity proper: problems with a short-sighted false-positive debate. Perspect Psychol Sci 7:661–669. https://doi.org/10.1177/1745691612462587
Xia J, Wishart DS (2016) Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr Protoc Bioinforma 55:14.10.1–14.10.91. https://doi.org/10.1002/cpbi.11
Fujita T, Hada T, Higashino K (1999) Origin of D- and L-pipecolic acid in human physiological fluids: a study of the catabolic mechanism to pipecolic acid using the lysine loading test. Clin Chim Acta Int J Clin Chem 287:145–156
Perera T, Young MR, Zhang Z et al (2015) Identification and monitoring of metabolite markers of dry bean consumption in parallel human and mouse studies. Mol Nutr Food Res 59:795–806. https://doi.org/10.1002/mnfr.201400847
Fukai K, Harada S, Iida M et al (2016) Metabolic Profiling of Total Physical Activity and Sedentary Behavior in Community-Dwelling Men. PLoS ONE 11:e0164877. https://doi.org/10.1371/journal.pone.0164877
Zhao Q, Shen H, Su K-J, et al (2018) A joint analysis of metabolomic profiles associated with muscle mass and strength in Caucasian women. Aging 10:2624–2635. https://doi.org/10.18632/aging.101574
Wikoff WR, Anfora AT, Liu J et al (2009) Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci 106:3698–3703. https://doi.org/10.1073/pnas.0812874106
de Mello VD, Paananen J, Lindström J et al (2017) Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish Diabetes Prevention Study. Sci Rep. https://doi.org/10.1038/srep46337
Tuomainen M, Lindström J, Lehtonen M et al (2018) Associations of serum indolepropionic acid, a gut microbiota metabolite, with type 2 diabetes and low-grade inflammation in high-risk individuals. Nutr Diabetes. https://doi.org/10.1038/s41387-018-0046-9
Jennis M, Cavanaugh CR, Leo GC et al (2018) Microbiota-derived tryptophan indoles increase after gastric bypass surgery and reduce intestinal permeability in vitro and in vivo. Neurogastroenterol Motil 30:e13178. https://doi.org/10.1111/nmo.13178
Abildgaard A, Elfving B, Hokland M et al (2018) The microbial metabolite indole-3-propionic acid improves glucose metabolism in rats, but does not affect behaviour. Arch Physiol Biochem 124:306–312. https://doi.org/10.1080/13813455.2017.1398262
Pallister T, Jennings A, Mohney RP et al (2016) Characterizing blood metabolomics profiles associated with self-reported food intakes in female twins. PLoS ONE 11:e0158568. https://doi.org/10.1371/journal.pone.0158568
Eggersdorfer M, Wyss A (2018) Carotenoids in human nutrition and health. Arch Biochem Biophys 652:18–26. https://doi.org/10.1016/j.abb.2018.06.001
Tomás-Barberán FA, Selma MV, Espín JC (2016) Interactions of gut microbiota with dietary polyphenols and consequences to human health. Curr Opin Clin Nutr Metab Care 19:471–476. https://doi.org/10.1097/MCO.0000000000000314
Steiber A, Kerner J, Hoppel CL (2004) Carnitine: a nutritional, biosynthetic, and functional perspective. Mol Aspects Med 25:455–473. https://doi.org/10.1016/j.mam.2004.06.006
Scalbert A, Brennan L, Manach C et al (2014) The food metabolome: a window over dietary exposure. AmJClinNutr 99:1286–1308
Shi L, Brunius C, Johansson I et al (2018) Plasma metabolites associated with healthy Nordic dietary indexes and risk of type 2 diabetes-a nested case-control study in a Swedish population. Am J Clin Nutr 108:564–575. https://doi.org/10.1093/ajcn/nqy145
Carayol M, Licaj I, Achaintre D et al (2015) Reliability of serum metabolites over a two-year period: a targeted metabolomic approach in fasting and non-fasting samples from EPIC. PLoSOne 10:e0135437
Floegel A, Drogan D, Wang-Sattler R et al (2011) Reliability of serum metabolite concentrations over a 4-month period using a targeted metabolomic approach. PLoS ONE 6:e21103
Acknowledgements
The authors thank Younes Esseddik, Frédéric Coffinieres, Thi Hong Van Duong, Paul Flanzy, Régis Gatibelza, Jagatjit Mohinder and Maithyly Sivapalan (computer scientists), Rachida Mehroug and Frédérique Ferrat (logistic assistants), Nathalie Arnault, Véronique Gourlet, PhD, Fabien Szabo, PhD, Julien Allegre, and Laurent Bourhis (data-manager/statisticians) and Cédric Agaesse (dietitian) for their technical contribution to the SU.VI.MAX study. We also thank all participants of the SU.VI.MAX study. This work was conducted in the framework of the French network for Nutrition And Cancer Research (NACRe network), www.inra.fr/nacre and received the NACRe Partnership Label. Metabolomic analyses were performed within the metaboHUB French infrastructure (ANR-INBS-0010).
Funding
This work was supported by the French National Cancer Institute [Grant number INCa_8085 for the project, and PhD Grant number INCa_11323 for L. Lecuyer]; the Federative Institute for Biomedical Research IFRB Paris 13; and the Cancéropôle Ile-de-France/Région Ile-de-France [PhD Grant for M. Deschasaux]. The funders had no role in the design, analysis, or writing of this article.
Author information
Authors and Affiliations
Contributions
The author’s responsibilities were as follow CM and MT: designed the research and supervised data interpretation; LL and CD conducted the research; SH, PG, MT, EKG: conducted the SU.VI.MAX cohort and provided essential data and samples; DC, ML, SD, BL and EPG: performed metabolomic analyses; LL, CD and PM: performed statistical analysis; MT, CM, MP: supervised statistical analysis; LL, MT, CD, CM and EPG: wrote the paper; all authors provided critical intellectual input for data interpretation, read and approved the final manuscript; CM and MT had primary responsibility for final content.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to disclose and the funders had no role in the design, analysis, or writing of this article.
Ethical approval
The study was conducted according to the guidelines laid down in the Declaration of Helsinki and approved by the Ethics Committee for Studies with Human Subjects of Paris-Cochin Hospital (CCPPRB 706/2364) and the ‘Commission Nationale de l’Informatique et des Libertés’ (CNIL 334641/907094). All subjects gave written informed consent to participate in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
394_2020_2177_MOESM1_ESM.docx
Supplementary file1 Loadings values of the a posteriori 3 dietary patterns derived by Principal Component Analysis (DOCX 27 kb)
394_2020_2177_MOESM4_ESM.docx
Supplementary file4 Identified metabolites associated with baseline clinical and biological parameters and nutrients intakes from Spearman correlation analyses (DOCX 58 kb)
394_2020_2177_MOESM5_ESM.docx
Supplementary file5 Associations between ions and the level of adherence to the French dietary recommendations, as measured by the a priori mPNNS-GS from multivariable conditional logistic regression (DOCX 50 kb)
394_2020_2177_MOESM6_ESM.docx
Supplementary file6 Associations between ions and components of the a priori mPNNS-GS from spearman correlation analyses (DOCX 363 kb)
394_2020_2177_MOESM7_ESM.docx
Supplementary file7 Associations between ions and a posteriori dietary patterns derived by principal component analysis from spearman correlation analyses (DOCX 96 kb)
Rights and permissions
About this article
Cite this article
Lécuyer, L., Dalle, C., Micheau, P. et al. Untargeted plasma metabolomic profiles associated with overall diet in women from the SU.VI.MAX cohort. Eur J Nutr 59, 3425–3439 (2020). https://doi.org/10.1007/s00394-020-02177-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00394-020-02177-5