Abstract
Human and other genomes encode tens of thousands of long noncoding RNAs (lncRNAs), the vast majority of which remain uncharacterised. High-throughput functional screening methods, notably those based on pooled CRISPR-Cas perturbations, promise to unlock the biological significance and biomedical potential of lncRNAs. Such screens are based on libraries of single guide RNAs (sgRNAs) whose design is critical for success. Few off-the-shelf libraries are presently available, and lncRNAs tend to have cell-type-specific expression profiles, meaning that library design remains in the hands of researchers. Here we introduce the topic of pooled CRISPR screens for lncRNAs and guide readers through the three key steps of library design: accurate annotation of transcript structures, curation of optimal candidate sets, and design of sgRNAs. This review is a starting point and reference for researchers seeking to design custom CRISPR screening libraries for lncRNAs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The number of annotated long noncoding RNA (lncRNA) genes has grown dramatically in the past decade thanks to next-generation sequencing (NGS). However, our ability to functionally characterise these genes has failed to keep pace, meaning that the vast majority of lncRNAs are of unknown biological or disease relevance (Ma et al. 2019). Into this gap has stepped CRISPR-Cas genome editing, and to a lesser extent other forms of pooled and arrayed screening, which together promise to mine this large unexplored genetic space and reveal new biological players and disease targets. The design of screening libraries is a foundation for such studies and is the focus of this Review.
Although the majority of lncRNAs remain uncharacterised, several hundred have already been linked to diseases or cell functions (Kung et al. 2013; Lekka and Hall 2018). Examples are MALAT1 and SAMMSON, which promote tumorigenesis in vitro and in vivo through relatively well-defined molecular mechanisms (Gutschner et al. 2013; Leucci et al. 2016). Somatic mutations and expression dysregulation of genes encoding these lncRNAs are observed in tumours, in addition to other clinical evidence such as expression correlation with patient survival (Vendramin et al. 2018; Chen et al. 2018; Vancura et al. 2021). This link to disease has raised considerable interest in lncRNAs as targets for precision RNA therapeutics (Arun et al. 2018; Esposito et al. 2019; Fathi 2020; Xiong et al. 2021).
Their exceedingly large numbers make it essential to screen for functional lncRNAs using high-throughput methods. Unfortunately, technologies developed for protein-coding genes (PCGs) face a number of barriers when applied to lncRNAs. First among these is that RNA interference (RNAi) is often ineffective for lncRNAs, possibly due to the latter’s relative enrichment in the nucleus (Maamar et al. 2013; Stojic et al. 2016). RNAi also generates large numbers of off-target hits (Smith et al. 2017) and generating new RNAi arrayed libraries is expensive and involves complex robotics equipment. Another hindrance arises from the relatively poor state of lncRNA gene annotation (Uszczynska-Ratajczak et al. 2018), which has hindered the development of off-the-shelf arrayed or pooled screening libraries.
These challenges have recently been overcome by rapid developments in gene perturbation technologies. Two effective perturbation methods are now available, which together map a path from initial screening to therapeutic use in patients. Clustered regularly interspaced short palindromic repeats (CRISPR) afford versatile and highly scalable perturbations in the laboratory via direct targeting of the lncRNA gene itself (Shalem et al. 2015). Antisense oligonucleotides (ASOs) achieve co-transcriptional degradation, representing both a powerful experimental tool and effective therapeutic, but at relatively low throughputs (Gutschner et al. 2013; Meng et al. 2015). Both CRISPR and ASOs are relatively low cost, practical, and have low off-target rates (Smith et al. 2017; Yoshida et al. 2019). Nonetheless, each method has drawbacks that must be mitigated. For example, CRISPR can be economically scaled to high throughputs, but wild-type (WT) CRISPR-Cas9 causes double strand breaks (DSBs) in DNA, whose toxicity can lead to unintended consequences (Chapman et al. 2012). This and other CRISPR approaches are highly sensitive to gene annotation quality. On the other hand, ASOs are relatively more costly to synthesise and are incompatible with pooled screening, which together have largely prevented their use at high throughputs. Nevertheless, these technologies, particularly CRISPR, open the door to economic high-throughput functional screening of lncRNAs.
All screening projects, including CRISPR, require the careful design of libraries of perturbation constructs. A critical input for such designs is accurate gene maps or annotations (Uszczynska-Ratajczak et al. 2018). The effectiveness of CRISPR perturbations is highly sensitive to correct targeting to gene’s TSS (Sanson et al. 2018). Unfortunately, annotations for lncRNAs tend to suffer from several issues, making them a constraint in CRISPR screens. We will discuss these issues in more detail and outline solutions to maximise annotation quality.
When performing high-throughput experiments for lncRNAs, a critical question to address is the following: “Which genes will we target?”. Only a minority of genes are likely to be candidates in a given biological system, not least because the cell model will only express a small fraction of the total “lncRNA-ome” (Jiang et al. 2016; Seifuddin et al. 2020). For example, Cabili et al. demonstrated that 78% of lncRNAs are expressed in a tissue-specific manner (Cabili et al. 2011). Depending on the type and aim of the screen, the pool of gene candidates can vary considerably. Also, the cost of the screen increases with the number of targets analysed. Therefore, the selection of the smallest optimal set of candidates is important for the economic and scientific success of a project.
Finally, the user must design perturbation constructs with optimised on-target efficacy and minimal off-target effects. In the case of CRISPR, this corresponds to single guide RNAs (sgRNAs). Our understanding of the sequence and genomic features determining these properties continues to evolve.
This Review aims to highlight the main aspects of an optimal high-throughput lncRNA screen and will cover these principle topics: evolution of lncRNA screen technologies, and the three steps of screening library design: gene annotation, candidate selection, and sgRNA design.
Functional screens for lncRNAs
Long noncoding RNAs at the frontier of biology and medicine
LncRNAs are defined as RNA transcripts longer than 200 nt that are not translated into proteins (Derrien et al. 2012). In comparison with the total number of PCGs, relatively stable at ~ 19,000 annotated genes (Frankish et al. 2019), the total number of lncRNA gene loci in humans is still under discussion with estimations ranging from 16,000 up to 140,000 (Ma et al. 2019; Frankish et al. 2019). Among these, just ~ 2000 have been functionally characterised in any detail (Ma et al. 2019).
Although an unknown number of lncRNAs may represent non-functional transcriptional noise (Palazzo and Lee 2015; Doolittle 2018) or be misannotated since they encode a small peptide (Ingolia et al. 2011), numerous studies have ascribed convincing roles and detailed molecular mechanisms to a core set of widely studied genes. For example, studies have demonstrated important roles for lncRNAs in regulation of embryonic development (Kung et al. 2013), DNA damage repair (Thapar 2018), chromatin remodelling and modifications (Marchese et al. 2017) among others. Similarly, lncRNAs play clear roles in human diseases, such as neuronal disorders (Sparber et al. 2019), cardiac diseases (Turton et al. 2019) and most notably cancer, where hundreds of lncRNAs have been functionally linked to tumorigenesis and cancer hallmarks (Schmitz et al. 2016; Schmitt and Chang 2017). In the above cases, lncRNAs have met the levels of evidence required for identifying PCG function, including in some cases, phenotypes in knockout animals (Adriaens et al. 2016; Wen et al. 2016; Akay et al. 2019; Gao et al. 2020). As a result, growing attention has gathered on the possibility of using lncRNAs as therapeutic targets to treat human diseases (Schmitt and Chang 2017; Chen et al. 2021).
LncRNAs present unique challenges to researchers. Their lack of encoded peptides means that the longstanding and effective functional prediction tools for proteins are ineffective for lncRNAs (Johnsson et al. 2014). Numerous attempts have been made to bioinformatically predict lncRNA functions; however, these usually rely on indirect evidence (for example, expression correlation) (Guo et al. 2013; Jiang et al. 2015; Pyfrom et al. 2019) whose predictive power is uncertain (Perron et al. 2017). Another widely employed source of evidence for functionality is evolutionary conservation (Chodroff et al. 2010; Carlevaro-Fita et al. 2019; Ruiz-Orera and Albà 2019), but here too lncRNAs are challenging: they tend to display low levels of evolutionary conservation at the sequence level, even for confidently functional cases like Cyrano (Ulitsky et al. 2011), while many others have no identifiable orthologues at all (Vendramin et al. 2018; Washietl et al. 2014; Necsulea et al. 2014; Hezroni et al. 2015). These considerations drive the search for innovative approaches to prioritise lncRNAs for functional screens.
Amongst the tens of thousands of remaining lncRNAs, there is a lively debate as to what proportion represent functional genes vs transcriptional noise (Palazzo and Lee 2015; Doolittle 2018). Regardless of the outcome, it is likely that thousands of novel genes with important biological and disease roles remain to be discovered. The enormous number of lncRNAs, coupled to our present lack of means of predicting their function a priori, makes high-throughput functional screens the only viable route to identifying the subset of functional genes.
Evolving tools for functional screening of lncRNAs
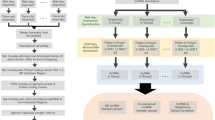
The large number of lncRNAs, coupled to our inability to predict their function, introduces the need for pooled functional screening approaches. Functional screening depends on two key factors: effective methods to perturb gene activity, and the degree to which such methods can be practically and economically scaled to high throughputs. The availability, or lack, of such techniques has dictated progress in lncRNA screening. Available perturbations fall into three principal types: RNA interference (RNAi) (effected by either small interfering RNAs or short hairpin RNAs); CRISPR-based perturbations; and ASOs (Fig. 1a–c). Here we introduce the principle perturbation methods for lncRNAs, then how they may be scaled to high throughputs by pooling.
Perturbation methods and mechanisms. Molecular mechanism of a RNA interference, b various CRISPR perturbations (CRISPR/Cas9 activity occurs in the nucleus, while CRISPR/Cas13 activity can occur in either the nucleus or the cytoplasm), and c antisense oligonucleotides (ASOs). d The main steps of a pooled CRISPR screen
Perturbation approaches: RNA interference
Early approaches to screen lncRNAs came from RNAi, which had a long history in PCG screening (Berns et al. 2004; Lord et al. 2008). RNAi depends on small (~ 22 bp) double-stranded RNAs that trigger degradation of complementary RNAs by the Argonaute family of proteins (Napoli et al. 1990; Fire et al. 1998; Cullen 2005). RNAi can be achieved by two distinct means: small interfering RNA (siRNA) and short hairpin RNA (shRNA). The two approaches differ in their delivery method (Fig. 1a), with important implications for screening. siRNA are chemically synthesised double-stranded oligonucleotides that must be delivered individually in an arrayed format, introducing the need for robotics and the generation of relatively expensive libraries (Rao et al. 2009). shRNAs are microRNA-like transcripts that are expressed as a single-stranded precursor, which folds into a hairpin structure and is recognised and processed into a double-stranded small RNA, similar to an siRNA (Elbashir et al. 2001; Caplen et al. 2001). shRNA genes may be delivered with a lentiviral plasmid, making them compatible with pooled screening (Sims et al. 2011). Given the topic of this Review, we here devote more space to shRNA; however, several important arrayed siRNA screens have been published (Whitehurst et al. 2007; Tiessen et al. 2019; Stojic et al. 2020).
shRNA has been used widely and successfully to screen PCGs, for example in Project Achilles (Tsherniak et al. 2017), although it is being rapidly supplanted by CRISPR (Bassik et al. 2009). The first pooled shRNA library for lncRNAs was designed to target 1280 intergenic mouse lncRNAs annotated in the ENSEMBL database (Lin et al. 2014). In a screen to identify lncRNAs involved in maintenance of pluripotency, the authors identified 20 hits, including TUNA. The size and focus of shRNA libraries can be adapted. For example, a larger library was designed for 3842 lncRNAs to identify those promoting proliferation of NIH3T3 mouse fibroblasts (Beermann et al. 2018). RNAi can also be adapted for in vivo experiments to study diseases. 120 lncRNAs were screened with a pooled shRNA library in a mouse model of acute myeloid leukemia, identifying 20 hits necessary for disease maintenance (Joaquina Delás et al. 2017).
Despite these successes, RNAi suffers from some notable drawbacks. First, RNAi perturbations often result in widespread unintended “off-target” repression of non-targeted genes (Smith et al. 2008). This is thought to occur as a result of the relatively short “seed” region through which RNAi target recognition takes place, resulting in large numbers of fortuitous matches in non-target genes (Birmingham et al. 2006; Sudbery et al. 2010). The outcome of this is observed phenotypic effects that arise independent of the intended target gene, i.e. false positives (Sudbery et al. 2010). The second principal drawback of RNAi is that it yields a variable and often low knockdown for lncRNAs (Lennox and Behlke 2016). The precise reasons for this remain unclear, and certainly many exceptions exist (Mondal et al. 2015; Gore-Panter et al. 2016), but it may be due to the preferential nuclear enrichment of lncRNAs (Carlevaro-Fita et al. 2019), whereas siRNA is more effective in the cytoplasm (Lennox and Behlke 2016; Zeng and Cullen 2002). A follow-on effect of this, is that it is suspected that even when successful lncRNA knockdown is observed, it may be the cytoplasmic RNA population that is preferentially affected, leaving nuclear activity intact (Maamar et al. 2013; Stojic et al. 2016). A more recent explanation came with the finding that siRNA requires translation to be effective, and hence only lncRNAs that are engaged by ribosomes will be impacted (Carlevaro-Fita et al. 2016; Biasini et al. 2021).
Thus, while a number of fruitful screens have been carried out for lncRNAs, both in pooled (Lin et al. 2014; Beermann et al. 2018; Joaquina Delás et al. 2017) and arrayed (Whitehurst et al. 2007; Tiessen et al. 2019; Stojic et al. 2020) formats, RNAi has not impacted the lncRNA field to the same extent as for PCGs, and researchers had to content themselves for many years with more conventional and low-throughput differential gene expression evidence as the starting point for identifying functional lncRNAs (Whitehurst et al. 2007; Lin et al. 2014).
Perturbation approaches: antisense oligonucleotides
A second perturbation approach worth mentioning is based on ASOs (Fire et al. 1998). While ASOs are not compatible with pooled screening, nevertheless they have become an indispensable tool for validating screen results. ASOs are short single-stranded oligonucleotides (13–25nt) that are chemically modified to achieve stability and potency (Dias and Stein 2002). ASOs hybridise by sequence complementarity to cellular RNAs and activate degradation by the enzyme RNase H (Crooke 2017) (Fig. 1c). ASOs display low off-target effects and are appropriate for use in humans for therapeutic applications (Crooke et al. 2021). Further advantages are their ability to be delivered into cells without the need of a delivery vehicle (“free uptake”), and particularly important for lncRNAs, they appear to degrade nascent RNAs in the process of transcription, thus accessing nuclear target populations (Pallarès-Albanell et al. 2019). However, due to the difficulty in designing effective on-target ASOs (typically around 40% are effective), the lower uptake efficiency when compared with vehicle mediated delivery methods (Stein et al. 2010; Hs et al. 2012) and the cost of their chemical synthesis, so far just one ASO screen for growth-modulating lncRNAs has been reported to date (Ramilowski et al. 2020).
Perturbation approaches: CRISPR
The advent of CRISPR genome editing has profoundly impacted the field of lncRNA functional genomics. For the first time, researchers have an effective tool that can be adapted to a variety of perturbations (repression, silencing, activation), can be targeted to the gene locus or the RNA product, displays reduced off-target effects, and most importantly, can be conveniently scaled to high throughputs (Sanson et al. 2018; Doench et al. 2016; Zhu et al. 2016; Diao et al. 2017; Gasperini et al. 2017).
CRISPR comprises an RNA:protein complex. The single guide RNA (sgRNA) consists of a 20 nt variable RNA sequence or “spacer”, which recognises by homology a specific genomic site followed by a protospacer adjacent motif (PAM) (Jinek et al. 2012; Mali et al. 2013; Cong et al. 2013; Sander and Joung 2014). The spacer is fused to an invariant structured “scaffold” that is recognised by the Cas9 protein. Researchers may target this complex to desired regions by simply identifying a PAM in that region, and designing the spacer sequence to recognise the adjacent 20mer. In turn, the Cas9 protein “cargo” may be engineered to perform various tasks at its destination, from DNA endonucleolytic cleavage in its wild-type form, to catalytically dead mutants (dCas9) fused to a growing array of effector domains (Fig. 1b) (Qi et al. 2013; Gilbert et al. 2013; Dominguez et al. 2016). Fusions carrying transcriptional inhibitor or activator domains form the basis for CRISPR inhibition (CRISPRi) and CRISPR activation (CRISPRa), respectively (Gilbert et al. 2013). This programmability enables CRISPR to be rapidly deployed for a wide range of desired perturbations (Doench et al. 2016). Because sgRNAs can be delivered by lentiviral vectors (Kosicki et al. 2018), CRISPR perturbations enable almost unlimited scalability in pooled screening format. Together these features make CRISPR a versatile and useful tool for discovering functional lncRNAs.
Nonetheless, CRISPR does present a number of hurdles that must be overcome. First of all, Cas9 is a bacterial protein and thus is highly immunogenic (Charlesworth et al. 2019). Induced Cas9 systems can help to mitigate this harmful effect in cells. WTCas9 nuclease activity results in double strand breaks (DSBs) that can cause genome rearrangements and cell death (Chapman et al. 2012; Kosicki et al. 2018; Leibowitz et al. 2021). The latter effect is stronger in cells expressing P53 (Bowden et al. 2020). The outcome is that sgRNAs may lead to non-specific apoptosis caused by the technique itself and not by the effect of the CRISPR modification. This must be addressed in screens by the careful design of phenotypically neutral controls: sgRNAs targeting intergenic regions give a better indication of background including DSB toxicity, rather than (often used) non-targeting controls, such as scrambled sequences, which will not cause DSBs and can lead to false positive hits (Aguirre et al. 2016; Haapaniemi et al. 2018). A second issue is off-targeting: while far lower than for shRNA (Smith et al. 2008), many sgRNAs do recognise non-targeted sites at non-zero frequency, resulting in off-target effects (Zhang et al. 2015). These effects can be largely avoided by careful sgRNA design using strict off-target filtering (Shalem et al. 2015). Higher concentrations of Cas9/sgRNA can lead to increased off-targets rates (Wu et al. 2014), therefore it is necessary to control for these concentration when performing in vivo experiments.
The first means of perturbing lncRNAs by CRISPR harnesses the ability of wild-type Cas9 to generate DSBs. This approach requires an understanding of the cellular processes that repair the resulting DSBs. The most prevalent pathway is non-homologous end joining (NHEJ), which is a non-templated method that repairs breaks but often introduces untemplated insertions and deletions (indels) at the repair site (Ceccaldi et al. 2016). These properties proved highly useful for knocking out PCGs, since sgRNAs targeted to open reading frames (ORFs) generate frameshift mutations that scramble the encoded peptide sequence (Shalem et al. 2015). Because, by definition, lncRNAs contain no encoded peptide, it is uncertain whether small indels are sufficient to impact lncRNA activity. Therefore, loss of function by CRISPR calls for more elaborate strategies. The most frequent approach is “CRISPR deletion” (CRISPR-del), where two wild-type CRISPR-Cas9 complexes are recruited to sites flanking a targeted genomic region (Aparicio-Prat et al. 2015). Simultaneous NHEJ gives rise to genomic deletion. Efficiency tends to lie in the range 40–60% of alleles (Gasperini et al. 2017; Aparicio-Prat et al. 2015; Kraft et al. 2015; Ran et al. 2013; Canver et al. 2014; Vidigal and Ventura 2015; Antoniani et al. 2018; Pulido-Quetglas et al. 2017), although often much less, and these rates broadly decline with the size of the deleted region(Canver et al. 2014).
CRISPR-del may be employed for lncRNA loss of function in several ways. The first and most obvious is by deletion of the entire gene body (Durruthy-Durruthy et al. 2015). However, this strategy entails several drawbacks. LncRNA genes can span several hundred kilobases. Such deletions tend to have low efficiency (Canver et al. 2014), and may well remove other overlapping functional elements, including PCGs and enhancers and thereby lead to false positive phenotypes. Removal of lncRNA TSS via targeted deletion of ~ 0.5 to 5 kb is a more practical alternative, by reducing the length of the deletion to a few hundreds to thousands of bases, increasing efficiency and uniformity, and decreasing the chance of deleting unrelated elements (Zhu et al. 2016; Pulido-Quetglas et al. 2017; Lavalou et al. 2019). Even effective deletions may not result in hoped for loss of gene expression: compensatory promoter activation has been reported in some cases (Lavalou et al. 2019). Given the deletion size mentioned, the TSS deletion strategy requires accuracy of lncRNA annotations at the 5′ end with a resolution of ~ 1 kb.
Other flavours of CRISPR can perturb lncRNA expression without permanently mutating DNA. By engineering appropriate fusion proteins with catalytically dead Cas9 (dCas9), one may achieve gene activation (CRISPRa) or inhibition (CRISPRi) (Liu et al. 2017; Horlbeck et al. 2016). Importantly, both these technologies require recruitment to a rather small window of ~ 200 bp with respect to the TSS, making them highly sensitive to accurate TSS annotation (Sanson et al. 2018). Resulting chromatin reorganisation of both methods can have and indirect effect on neighbouring genes. CRISPRa mechanism is capable of open the chromatin and allows transcription machinery to access genes located nearby the targeted region increasing their expression. Similarly, indirect reduction in gene expression can be observed when targeting genes with CRISPRi (Horlbeck et al. 2016; Groner et al. 2010). Researchers should therefore validate the results obtained by these methods analysing any unintended changes in expression of nearby genes. These approaches avoid issues of DSB toxicity, while having the additional benefit of being compatible with a variety of inducible systems, affording the researcher temporal control over gene perturbations (Sun et al. 2019).
Pooled screening
Genetic screens are a powerful method to test the effect of gene perturbations in a high-throughput way (Sanson et al. 2018; Doench et al. 2016; Zhu et al. 2016; Liu et al. 2017). Screens in cultured cells can be performed in two formats: arrayed and pooled. Arrayed screens apply a single perturbation to multiple cells in one well. They require robotics equipment, due to the large number of wells involved, and they require synthesis of many individual perturbation reagents (siRNAs or sgRNAs) (Lord et al. 2008; Whitehurst et al. 2007). Screen results are read out from each individual well, and as a result are relatively unconstrained in terms of the phenotypic features that can be measured, extending to microscopy and image analysis (Stojic et al. 2020).
Pooled screens, in contrast, involve introducing a mixed pool of perturbation constructs into a single cell population (Fig. 1d). Libraries are synthesised as a mixture using increasingly inexpensive oligonucleotide “megasynthesis” (Doench 2017), and delivered with genomically integrating lentiviruses (Sanson et al. 2018). Viruses are usually applied at low titres (multiplicity of infection, MOI, ~ 0.3), so that every cell in the population carries one perturbation. A selection is applied in order to isolate two or more cell populations with different phenotypes. Genomically integrated perturbation sequences, usually sgRNAs, are then used as barcodes to determine the differences in library composition between cell populations of different phenotypes, and hence infer functional lncRNAs contributing to said phenotypes (Zhu et al. 2019; Boettcher et al. 2019). This highlights the key constraint of pooled screening: phenotypic readouts are restricted to those which can be sorted in some way (Sanson et al. 2018; Shalem et al. 2015). These include cell fitness/proliferation, fluorescence, survival in response to insult, or migration(Sanson et al. 2018; Zhu et al. 2016; Shalem et al. 2015; Liu et al. 2018), but rules out imaging-based readouts.
Even with the requirements of pooled screens—i.e. phenotypic selection, next generation sequencing, and deconvolution of the data to determine perturbation abundances—the benefits these screens provide compared to arrayed screens are significant. In pooled screens, libraries can be created, delivered to cells and analysed as a single sample, considerably reducing the cost and hands-on time. This also avoids the capital investment and training required for robotics necessary for arrayed screening. The fact that only a single sample has to be analysed, helps to reduce batch effects and increases the statistical power of the analysis, since all perturbations, tests and controls, are treated with the same exact conditions. These advantages have led to growing adoption of pooled CRISPR screens.
A key requirement of pooled screens is the screening library. Libraries targeting all or subsets of PCGs are rapidly growing in quality, and are available from multiple suppliers (Doench et al. 2016). In contrast, few such resources are presently available for lncRNAs, due to a number of factors. Firstly, the number and quality of lncRNA annotations increases so rapidly that libraries rapidly become obsolete. Secondly, lncRNAs have highly cell-type-specific expression profiles, meaning that available libraries designed for a given purpose, may not cover a useful proportion of targets in a different biological assay or cellular background. These factors mean that researchers are likely to have to design custom lncRNA screening libraries for the immediate future. Provide a uniform guideline is the purpose of the present Review. The process of designing a screening library can be broken into three principle steps: gene annotation, candidate selection, and sgRNA design (Fig. 2a). These steps are explained in more detail in the following sections.
Accurate transcript annotations
Gene perturbation, particularly by CRISPR, depends critically on recruiting Cas9 to a narrowly defined window around the TSS (Sanson et al. 2018). Consequently, accurate maps of gene and transcript structures are essential for functional screening (Sanson et al. 2018; Bergadà-Pijuan et al. 2020). These maps are referred to as annotations, and specify the exact location of gene’s constituent transcripts, introns and exons (Harrow et al. 2006). Most importantly in the present context, annotations record the expected location of TSSs, being simply the start position of the first exon for the transcript(s) comprising a gene (Fig. 2b).
Despite their importance, lncRNA annotations remain an imperfect reflection of the underlying biological reality, and are best regarded as work in progress, provided by several different sources and created with different approaches (Uszczynska-Ratajczak et al. 2018). GENCODE, for example, provides lncRNA annotations for ENSEMBL and is a mixture of manual and high-quality experimental annotations, which ensures a good quality but relatively small size and incomplete coverage for many cell types (Uszczynska-Ratajczak et al. 2018; Derrien et al. 2012; Lagarde et al. 2017). This and the other principle manually curated resource, RefSeq, have formed the basis for several shRNA and CRISPR screen designs for human and mouse genomes (Lin et al. 2014; Beermann et al. 2018; Zhu et al. 2016). A more complete and detailed lists of resources for lncRNA annotations can be found in two reviews: Uszczynska-Ratajczak et al. (Uszczynska-Ratajczak et al. 2018) and Richard et al. (Charles Richard and Eichhorn 2018).
An important drawback of the above public annotations, is that they are not comprehensive—they omit many genuine lncRNAs (Uszczynska-Ratajczak et al. 2018). This may occur due to the distinct annotation protocols and criteria employed. However, another important cause is the fact that annotations are based on published transcriptomic resources, or from focussed studies in a small number of cell types (Lagarde et al. 2017). Thus, the lncRNAs they contain are biassed towards those expressed in widely studied cell lines and organs. This will impact researchers who wish to perform a screen in any cell model that is not well represented in the above datasets.
Two solutions are available to the researcher to address this lack of annotation comprehensiveness in their model of interest. The first is to merge several public annotations into a single, larger and more comprehensive one. Several software packages are available for this (Trapnell et al. 2010; Pertea et al. 2015). A second, more time-consuming but more effective approach, is to create a custom annotation through transcriptome assembly (Grabherr et al. 2011; Kovaka et al. 2019; Hölzer and Marz 2019). By using RNA-sequencing data from the cell model of interest, a new “assembly” of transcriptome annotation can be built bioinformatically (Liu et al. 2020). The advantage here is that the assembly reflects the transcriptome in the cells where the screen is to be performed. Thus, it might contain many novel and cell specifically expressed lncRNAs that are missing in public annotations (Roberts et al. 2011). Novel assemblies are usually further merged with public assemblies for extra confidence (Joaquina Delás et al. 2017; Liu et al. 2017). Transcriptome assemblies are algorithmically predicted from short RNA-sequencing fragments and therefore they might not be 100% accurate. In future, this gap can be mitigated by using long read RNAseq data, which captures the full sequence of lncRNA transcripts and the assembly step will not be necessary (Lagarde et al. 2017).
A second key feature of annotations is their completeness—or whether they accurately record the location of the TSS (Uszczynska-Ratajczak et al. 2018). LncRNAs can present multiple TSSs and correct identification is crucial (Mattioli et al. 2019; Kindgren et al. 2018). As mentioned before, CRISPR perturbations depend on recruitment to a small window around the TSS, meaning that even minor inaccuracies in TSS annotation may result in false negative results. Unfortunately, lncRNA annotations are poor at correctly recording TSS locations, as defined by gold-standard evidence from Cap Analysis of Gene Expression (CAGE), a sensitive method to map 5′ ends of transcripts (Uszczynska-Ratajczak et al. 2018; Hon et al. 2017). Although transcriptome assemblies have particularly poor performance at identifying TSS (Lagarde et al. 2017) the FANTOM group has accurately re-annotated lncRNA TSSs from multiple transcripts collections by including CAGE analysis into the analysis (Hon et al. 2017).
Both of these issues with lncRNA annotations, missing genes or incompleteness at 5′ end, will ultimately result in false negatives. This was demonstrated recently by reanalysis of published CRISPRi screens, where it was found that lncRNAs are significantly less likely to be hits when their TSS is inaccurately annotated (as judged by CAGE data) (Bergadà-Pijuan et al. 2020).
Another key variable for researchers is the species under study. Despite their drawbacks, lncRNA annotations in human and mouse are far more advanced than other model organisms (Sundaram et al. 2017). Less is known about lncRNA populations in non-model species, although we have no reason to believe they are any less important or numerous. Researchers working on non-model species will, given their lack of lncRNA annotations, have to rely even more on transcriptome assemblies for library design.
Narrowing down the best candidates
The number of genes that can be included in a screen is limited by cost and other practical parameters. A typical CRISPR screen requires multiple sgRNAs per target gene (usually ~ 10), a coverage of 100–1000 individual cells per sgRNA sequence, and 100 s of NGS reads per sgRNA (Sanson et al. 2018; Doench 2017). Therefore, materials cost increases with the number of candidates tested. Fortunately, it is not necessary to screen the entire population of 100,000 + annotated lncRNAs (Fang et al. 2018), because only a small subset are present in a given cell model. Thus, the second step of library design involves filtering to focus on a reduced set of candidate lncRNAs that are most likely to contain screen hits. More so than the other two steps, this one is most specific to the particular biological system under study and requires the greatest amount of user discretion.
Several filtering methods can be applied in order to enrich the final list of candidates for likely hits (Fig. 3a). The primary and most obvious filter is expression in the cells of interest. In principle, only expressed transcripts should be biologically active, and the majority of silent lncRNA genes can be omitted. Thus, it will be necessary to quantify specific RNAseq data from the screen model to select those lncRNAs expressed. For example, in Liu et al. only ENSEMBL lncRNAs expressed in the cell lines used in the study were included in the screen (Liu et al. 2018). Due to the low expression levels of lncRNAs, thresholds as low as 0.1 transcripts per million (TPM) can be required to not miss any relevant lncRNAs, especially given the exceedingly low expression observed for some functional lncRNAs (Seiler et al. 2017). This step alone will substantially narrow the candidate set, and indeed may alone be sufficient to reach the desired library size.
Another important consideration for candidate selection is gene copy number. When the goal is a complete knockout, it will be more challenging to achieve for genes present at > 2 copies per cell. Furthermore, targeting these genes with CRISPR will generate multiple DSBs, increasing the likelihood of non-specific toxicity to the cell (Aguirre et al. 2016). Information for the gene copy number in multiple cell lines can be obtained from the Cancer Cell Line Encyclopedia (CCLE; https://sites.broadinstitute.org/ccle/). These considerations are further complicated by the fact that oncogenes are frequently amplified in tumours, meaning that phenotypic effects of targeting oncogenic lncRNAs may be a mixture of both specific and non-specific effects. To our knowledge, this issue remains to be satisfactorily resolved, apart from careful validation by ASOs or other DSB-independent perturbations. Differential expression can also be used as a method for selection. For example, when screening for lncRNAs involved in cancer development, tumor samples can be compared against its healthy counterpart to find tumour-upregulated lncRNAs (Zhu et al. 2016).
Some of the filters will be already imposed by the screen method itself. For example, if we use a CRISPR deletion approach, only intergenic (non-PCG overlapping) lncRNAs might be targeted, so as to avoid perturbation of a nearby PCG. In this case a minimum distance from the TSS of lncRNAs to the nearest PCG can be applied as a filter.
Many lncRNAs have been associated with diseases. Several online databases have compiled such lncRNAs, and may be used as a valuable filter for candidate selection (Vancura et al. 2021; Bao et al. 2019; Wang et al. 2019; Zhao et al. 2020). The drawback of this approach is that it will omit novel lncRNAs from transcriptome assemblies. Other useful evidence for lncRNA function in disease may come from germline variants lying nearby (Giral et al. 2018; Aznaourova et al. 2020) (or better, that are also quantitative expression trait loci or eQTLs) (Goede et al. 2021). Similarly, somatic single nucleotide variants or copy number variants are also important evidence for prioritising cancer lncRNAs (Lanzós et al. 2017; Minotti et al. 2018; Gao et al. 2019). The latter datasets (with the exception of eQTLs) have the added benefit of being compatible with novel transcriptome assemblies.
LncRNAs are evolutionarily less conserved than PCGs (Uszczynska-Ratajczak et al. 2018). However, the conservation of their exon structure and expression pattern in different developmental stages across related species is important evidence for functionality (Chodroff et al. 2010; Hezroni et al. 2015; Sarropoulos et al. 2019; Carlevaro-Fita et al. 2020). Although conservation is not a limitation (Ruan et al. 2020), the presence of orthologues in other species can be used as a filter for screen candidates.
After filtering and selecting the optimal lncRNA candidates only one step remains: the design of an optimised library of perturbation constructs.
Designing sgRNA libraries
CRISPR perturbation efficiency is directly linked to sgRNA design. Improved designs will increase the performance of the screen and avoid false negatives. The 20 nt sgRNA spacer sequence will dictate the on-target activity and the number of possible off-target regions (Doench et al. 2016; Abadi et al. 2017; Liu et al. 2020). Although sgRNA sequence itself and folding stability are key to increase the efficiency for the on-target region, orientation of the guide in relation to the target gene has also been reported to impact sgRNA efficiency (Wang et al. 2014). Various algorithms and tools are available to design and calculate on-target efficiency of sgRNAs from query sequences or gene IDs (Doench et al. 2016; Horlbeck et al. 2016; Xu et al. 2015; Wong et al. 2015; Concordet and Haeussler 2018). One of these algorithms also account for CRISPRi/a designs (Sanson et al. 2018; Doench et al. 2016). Of relevance for lncRNAs screens, tools for paired sgRNA designs are also available (Pulido-Quetglas et al. 2017; Perez et al. 2017). An important drawback of tools accepting only gene IDs, is that sgRNAs can only be designed for known genes, leaving novel genes and non-genic regions untargetable. Some tools only provide sgRNA designs for one or a limited number of targets at a time, which makes them unsuitable for scaling up to high throughputs, while others can provide designs for an unlimited number of target regions (Pulido-Quetglas et al. 2017) (https://portals.broadinstitute.org/gppx/crispick/public). A summary of these tools can be found in Table 1.
Genomic regions with complementarity to the sgRNAs can cause undesired off-target effects. Off-target regions can tolerate mismatches, particularly when they fall more distal to the PAM end of the sgRNA-DNA hybrid (Hsu et al. 2016). Removing sgRNAs with potential off-target matches is routinely performed in library designs (Doench et al. 2016). Several online tools are also available to find off-target regions and calculate their scores (Doench et al. 2016; Bae et al. 2014; Stemmer et al. 2015). Different scoring algorithms will rank sgRNAs differently, thus, concordance between predicted and measured activity of the guide can vary (Labuhn et al. 2018). To mitigate the fluctuation of efficiency and to increase statistical power, it is common practice to design several sgRNA (4–10) per target region (Bodapati et al. 2020) (Fig. 4). This number can be reduced to two with optimal sgRNAs targeting known essential control genes (Wong et al. 2015). While broadly used genome-wide Cas9 libraries targeting PCGs have on average 105 sgRNAs, this number is halved for lncRNA targeting CRISPR screens.
Not only the characteristics of the sgRNA are important but also the location of the on-target regions. For example, in CRISPR deletion screens, the distance between the two sgRNAs will have an impact on the efficiency. Although large deletions have been achieved (Mizuno-Iijima et al. 2020) efficiency decreases for deletions larger than 0.5—5 k bps (Fig. 4a) (Zhu et al. 2016; Canver et al. 2014; Han et al. 2014; Zheng et al. 2014). CRISPRi and CRISPRa efficiency also depends on the distance of the on-target region to the targeted TSS. Optimal sgRNA design ranges for this approaches are extremely narrow, lying between + 25 to + 75nts downstream of the TSS for CRISPRi and − 150 to − 75 nts upstream of the TSS for CRISPRa (Sanson et al. 2018; Bergadà-Pijuan et al. 2020; Radzisheuskaya et al. 2016) (Fig. 4b and c). CAGE data can be used to select optimal transcript TSSs to optimise CRISPRi/a designs (Sanson et al. 2018).
Positive and negative controls are crucial to properly analyse the performance of the library and to measure the CRISPR perturbation effect. A total of at least 300 sgRNAs targeting positive control genes are needed to effectively control the false discovery rate (Bodapati et al. 2020). Genes known to influence the screening phenotype are typically used as positive controls (Aguirre et al. 2016; Haapaniemi et al. 2018). Essential genes, such as those encoding ribosomal proteins, or growth-promoting genes, are frequently employed as positive controls in CRISPR screens based on cell fitness/proliferation (Zhu et al. 2016; Liu et al. 2018), as their sgRNAs should disappear or “drop out” in the final population of cells. A minimum of three sgRNAs should be used to target these controls. Negative controls (sometimes referred as neutral controls) are not expected to influence phenotype and are used as a reference with which to identify screen hits. Intergenic regions (Zhu et al. 2016) or the Adeno-Associated Virus Integration Site 1 (AAVS1) where deletions have been proved non-deleterious (Smith et al. 2008; Chu et al. 2015; Hayashi et al. 2020) are good choices for design of negative control sgRNAs (Zhu et al. 2016; Liu et al. 2018). In experiments with wild-type Cas9, we recommend the use of targeting negative controls (i.e. that target a non-functional genomic region) rather than non-targeting controls (i.e. containing a spacer with no genomic match), since the former more accurately model the non-specific toxicity arising from DSBs.
Outlook
The discovery of functional lncRNAs has been revolutionised by pooled screening technology, particularly that implemented with CRISPR and its variants. CRISPR screening is capable of functionally interrogating thousands of lncRNAs in a single experiment, without the overheads associated with arrayed screening. Its favourable performance across multiple features (reduced off-targets, high on-target efficiency, flexible delivery method and high programmability) has led to its rapid adoption over RNAi-based approaches. As the volume of CRISPR data increases, further improvements are likely in aspects such as sgRNA on-target and off-target activity.
Space constraints meant that we could not discuss an upcoming variation of CRISPR screening based on direct RNA perturbation with Cas13 and other enzymes (Anton et al. 2018; Cox et al. 2017; Abudayyeh et al. 2017; Xu et al. 2020). Instead of targeting the gene, RNA-targeted CRISPR directly destabilises or otherwise perturbs the RNA transcript itself. Although it is still in development, several publications have already demonstrated its efficiency in different organisms (Yang et al. 2019; Kushawah et al. 2020; Huynh et al. 2020). Similar to CRISPRi/a, Cas13 can be converted into a programmable RNA binding platform by mutating its catalytic site (dCas13) and fusing it to a catalytic enzyme with desired activities. In this way, dCas13 could be use, for example, as a tool for live cell RNA imaging (Palaz et al. 2021). This may be a promising option for gene therapy applications, where DNA mutation is undesirable (Anton et al. 2020).
The practicality and versatility of CRISPR screening makes it capable of identifying lncRNAs mediating a wide variety of cellular processes in healthy and diseased biological contexts. We expect that as annotations improve and screen components become standardised, this approach will become increasingly widely used to identify molecular components and therapeutic targets among the tens of thousands of uncharacterised lncRNA genes.
References
Abadi S, Yan WX, Amar D, Mayrose I (2017) A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comput Biol 13(10):e1005807. https://doi.org/10.1371/journal.pcbi.1005807
Abudayyeh OO, Gootenberg JS, Essletzbichler P et al (2017) RNA targeting with CRISPR–Cas13. Nature. https://doi.org/10.1038/nature24049
Adriaens C, Standaert L, Barra J et al (2016) P53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med 22(8):861–868. https://doi.org/10.1038/nm.4135
Aguirre AJ, Meyers RM, Weir BA et al (2016) Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov 6(8):914–929. https://doi.org/10.1158/2159-8290.CD-16-0154
Akay A, Jordan D, Navarro IC et al (2019) Identification of functional long non-coding RNAs in C. elegans. BMC Biol 17(1):1–14. https://doi.org/10.1186/s12915-019-0635-7
Anton T, Karg E, Bultmann S (2018) Applications of the CRISPR/Cas system beyond gene editing. Biol Methods Protoc. https://doi.org/10.1093/biomethods/bpy002
Antoniani C, Meneghini V, Lattanzi A et al (2018) Induction of fetal hemoglobin synthesis by CRISPR/Cas9-mediated editing of the human b-globin locus. Blood 131(17):1960–1973. https://doi.org/10.1182/blood-2017-10-811505
Aparicio-Prat E, Arnan C, Sala I, Bosch N, Guigó R, Johnson R (2015) DECKO: single-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genomics 16(1):1–15. https://doi.org/10.1186/s12864-015-2086-z
Arun G, Diermeier SD, Spector DL (2018) Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med 24(3):257–277. https://doi.org/10.1016/j.molmed.2018.01.001
Aznaourova M, Schmerer N, Schmeck B, Schulte LN (2020) Disease-causing mutations and rearrangements in long non-coding RNA gene loci. Front Genet 11:1485. https://doi.org/10.3389/fgene.2020.527484
Bae S, Park J, Kim J-S (2014) Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30(10):1473–1475. https://doi.org/10.1093/bioinformatics/btu048
Bao Z, Yang Z, Huang Z, Zhou Y, Cui Q, Dong D (2019) LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res 47:D1034–D1037. https://doi.org/10.1093/nar/gky905
Bassik MC, Lebbink RJ, Churchman LS et al (2009) Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat Methods 6(6):443–445. https://doi.org/10.1038/nmeth.1330
Beermann J, Kirste D, Iwanov K et al (2018) A large shRNA library approach identifies lncRNA Ntep as an essential regulator of cell proliferation. Cell Death Differ 25(2):307–318. https://doi.org/10.1038/cdd.2017.158
Bergadà-Pijuan J, Pulido-Quetglas C, Vancura A, Johnson R (2020) CASPR, an analysis pipeline for single and paired guide RNA CRISPR screens, reveals optimal target selection for long non-coding RNAs. Bioinformatics 36(6):1673–1680. https://doi.org/10.1093/bioinformatics/btz811
Berns K, Hijmans EM, Mullenders J et al (2004) A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428(6981):431–437. https://doi.org/10.1038/nature02371
Biasini A, Abdulkarim B, Pretis S et al (2021) Translation is required for miRNA-dependent decay of endogenous transcripts. EMBO J 40(3):e104569. https://doi.org/10.15252/embj.2020104569
Birmingham A, Anderson EM, Reynolds A et al (2006) 3’ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods 3(3):199–204. https://doi.org/10.1038/nmeth854
Bodapati S, Daley TP, Lin X, Zou J, Qi LS (2020) A benchmark of algorithms for the analysis of pooled CRISPR screens. Genome Biol 21(1):1–13. https://doi.org/10.1186/s13059-020-01972-x
Boettcher M, Covarrubias S, Biton A et al (2019) Tracing cellular heterogeneity in pooled genetic screens via multi-level barcoding. BMC Genomics 20(1):1–9. https://doi.org/10.1186/s12864-019-5480-0
Bowden AR, Morales-Juarez DA, Sczaniecka-Clift M et al (2020) Parallel crispr-cas9 screens clarify impacts of p53 on screen performance. Elife 9:1. https://doi.org/10.7554/eLife.55325
Cabili MN, Trapnell C, Goff L et al (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25(18):1915–1927. https://doi.org/10.1101/GAD.17446611
Canver MC, Bauer DE, Dass A et al (2014) Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/cas9 nuclease system in mammalian cells. J Biol Chem 289(31):21312–21324. https://doi.org/10.1074/jbc.M114.564625
Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA (2001) Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci USA 98(17):9742–9747. https://doi.org/10.1073/pnas.171251798
Carlevaro-Fita J, Rahim A, Guigó R, Vardy LA, Johnson R (2016) Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 22(6):867–882. https://doi.org/10.1261/rna.053561.115
Carlevaro-Fita J, Polidori T, Das M, Navarro C, Zoller TI, Johnson R (2019) Ancient exapted transposable elements promote nuclear enrichment of human long noncoding RNAs. Genome Res 29(2):208–222. https://doi.org/10.1101/gr.229922.117
Carlevaro-Fita J, Lanzós A, Feuerbach L et al (2020) Cancer LncRNA census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis. Commun Biol 3(1):1–16. https://doi.org/10.1038/s42003-019-0741-7
Ceccaldi R, Rondinelli B, D’Andrea AD (2016) Repair pathway choices and consequences at the double-strand break. Trends Cell Biol 26(1):52–64. https://doi.org/10.1016/j.tcb.2015.07.009
Chapman JR, Taylor MRG, Boulton SJ (2012) Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47(4):497–510. https://doi.org/10.1016/j.molcel.2012.07.029
Charles Richard JL, Eichhorn PJA (2018) Platforms for investigating LncRNA functions. SLAS Technol 23(6):493–506. https://doi.org/10.1177/2472630318780639
Charlesworth CT, Deshpande PS, Dever DP et al (2019) Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med 25(2):249. https://doi.org/10.1038/S41591-018-0326-X
Chen Y, Huang W, Sun W et al (2018) LncRNA MALAT1 promotes cancer metastasis in osteosarcoma via activation of the PI3K-Akt signaling pathway. Cell Physiol Biochem 51:1313–1326. https://doi.org/10.1159/000495550
Chen Y, Li Z, Chen X, Zhang S (2021) Long non-coding RNAs: from disease code to drug role. Acta Pharm Sin b 11(2):340–354. https://doi.org/10.1016/j.apsb.2020.10.001
Chodroff RA, Goodstadt L, Sirey TM et al (2010) Long noncoding RNA genes: caaonservation of sequence and brain expression among diverse amniotes. Genome Biol 11(7):1–16. https://doi.org/10.1186/gb-2010-11-7-r72
Chu VT, Weber T, Wefers B et al (2015) Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33(5):543–548. https://doi.org/10.1038/nbt.3198
Concordet JP, Haeussler M (2018) CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res 46(W1):W242–W245. https://doi.org/10.1093/nar/gky354
Cong L, Ran FA, Cox D et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121):819–823. https://doi.org/10.1126/science.1231143
Cox DBT, Gootenberg JS, Abudayyeh OO et al (2017) RNA editing with CRISPR-Cas13. Science 358(6366):1019–1027. https://doi.org/10.1126/science.aaq0180
Crooke ST (2017) Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther 27(2):70–77. https://doi.org/10.1089/nat.2016.0656
Crooke ST, Baker BF, Crooke RM, Liang X (2021) Antisense technology: an overview and prospectus. Nat Rev Drug Discov 20(6):427–453. https://doi.org/10.1038/s41573-021-00162-z
Cullen BR (2005) RNAi the natural way. Nat Genet 37(11):1163–1165. https://doi.org/10.1038/ng1105-1163
de Goede OM, Nachun DC, Ferraro NM et al (2021) Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell 184(10):2633-2648.e19. https://doi.org/10.1016/j.cell.2021.03.050
Derrien T, Johnson R, Bussotti G et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22(9):1775–1789. https://doi.org/10.1101/gr.132159.111
Diao Y, Fang R, Li B et al (2017) A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat Methods. https://doi.org/10.1038/nmeth.4264
Dias N, Stein CA (2002) Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Therap 1:347–355
Doench JG (2017) Am I ready for CRISPR? A user’s guide to genetic screens. Nat Rev Genet 19(2):67–80. https://doi.org/10.1038/nrg.2017.97
Doench JG, Fusi N, Sullender M et al (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34(2):184–191. https://doi.org/10.1038/nbt.3437
Dominguez AA, Lim WA, Qi LS (2016) Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol 17(1):5–15. https://doi.org/10.1038/nrm.2015.2
Doolittle WF (2018) We simply cannot go on being so vague about “function.” Genome Biol 19(1):1–3. https://doi.org/10.1186/s13059-018-1600-4
Durruthy-Durruthy J, Sebastiano V, Wossidlo M et al (2015) The primate-specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nat Genet 48(1):44–52. https://doi.org/10.1038/ng.3449
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411(6836):494–498. https://doi.org/10.1038/35078107
Esposito R, Bosch N, Lanzós A, Polidori T, Pulido-Quetglas C, Johnson R (2019) Hacking the cancer genome: profiling therapeutically actionable long non-coding RNAs using CRISPR-Cas9 screening. Cancer Cell 35(4):545–557. https://doi.org/10.1016/j.ccell.2019.01.019
Fang S, Zhang L, Guo J et al (2018) NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res 46(D1):D308–D314. https://doi.org/10.1093/nar/gkx1107
Fathi DB (2020) Strategies to target long non-coding RNAs in cancer treatment: progress and challenges. Egypt J Med Hum Genet 21(1):1–15. https://doi.org/10.1186/s43042-020-00074-4
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391(6669):806–811. https://doi.org/10.1038/35888
Frankish A, Diekhans M, Ferreira AM et al (2019) GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47(D1):D766–D773. https://doi.org/10.1093/nar/gky955
Gao Y, Li X, Zhi H et al (2019) Comprehensive characterization of somatic mutations impacting lncRNA expression for pan-cancer. Mol Ther - Nucleic Acids 18:66–79. https://doi.org/10.1016/j.omtn.2019.08.004
Gao F, Cai Y, Kapranov P, Xu D (2020) Reverse-genetics studies of lncRNAs-what we have learnt and paths forward. Genome Biol 21(1):1–23. https://doi.org/10.1186/s13059-020-01994-5
Gasperini M, Findlay GM, McKenna A et al (2017) CRISPR/Cas9-mediated scanning for regulatory elements required for HPRT1 expression via thousands of large, programmed genomic deletions. Am J Hum Genet 101(2):192–205. https://doi.org/10.1016/j.ajhg.2017.06.010
Gilbert LA, Larson MH, Morsut L et al (2013) XCRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154(2):442. https://doi.org/10.1016/j.cell.2013.06.044
Giral H, Landmesser U, Kratzer A (2018) Into the wild: GWAS exploration of non-coding RNAs. Front Cardiovasc Med 5:181. https://doi.org/10.3389/fcvm.2018.00181
Gore-Panter SR, Hsu J, Barnard J et al (2016) PANCR, the PITX2 adjacent noncoding RNA, is expressed in human left atria and regulates PITX2c Expression. Circ Arrhythmia Electrophysiol. https://doi.org/10.1161/CIRCEP.115.003197
Grabherr MG, Haas BJ, Yassour M et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644–652. https://doi.org/10.1038/nbt.1883
Groner AC, Meylan S, Ciuffi A et al (2010) KRAB–zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet 6(3):1000869. https://doi.org/10.1371/JOURNAL.PGEN.1000869
Guo X, Gao L, Liao Q et al (2013) Long non-coding RNAs function annotation: a global prediction method based on bi-colored networks. Nucleic Acids Res 41(2):e35–e35. https://doi.org/10.1093/nar/gks967
Gutschner T, Hämmerle M, Eißmann M et al (2013) The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 73(3):1180–1189. https://doi.org/10.1158/0008-5472.CAN-12-2850
Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J (2018) CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 24(7):927–930. https://doi.org/10.1038/s41591-018-0049-z
Han J, Zhang J, Chen L et al (2014) Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol 11(7):829–835. https://doi.org/10.4161/rna.29624
Harrow J, Denoeud F, Frankish A et al (2006) GENCODE: producing a reference annotation for ENCODE. Genome Biol 7:4. https://doi.org/10.1186/gb-2006-7-s1-s4
Hayashi H, Kubo Y, Izumida M, Matsuyama T (2020) Efficient viral delivery of Cas9 into human safe harbor. Sci Rep 10(1):1–14. https://doi.org/10.1038/s41598-020-78450-8
Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I (2015) Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep 11(7):1110–1122. https://doi.org/10.1016/j.celrep.2015.04.023
Hölzer M, Marz M (2019) De novo transcriptome assembly: a comprehensive cross-species comparison of short-read RNA-Seq assemblers. Gigascience 8(5):1–16. https://doi.org/10.1093/gigascience/giz039
Hon C-C, Ramilowski JA, Harshbarger J et al (2017) An atlas of human long non-coding RNAs with accurate 5′ ends. Res Cent Biotechnol RAS 6009(7644):199–204. https://doi.org/10.1038/nature21374
Horlbeck MA, Gilbert LA, Villalta JE et al (2016) Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife 5:e19760. https://doi.org/10.7554/eLife.19760
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ (2014) DNA targeting specificity of RNA-guided Cas9 nucleases Patrick. Nat Biotechnol 31(9):827–832. https://doi.org/10.1038/nbt.2647.DNA
Huynh N, Depner N, Larson R, King-Jones K (2020) A versatile toolkit for CRISPR-Cas13-based RNA manipulation in Drosophila. Genome Biol 21(1):1–29. https://doi.org/10.1186/s13059-020-02193-y
Ingolia NT, Lareau LF, Weissman JS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147(4):789–802. https://doi.org/10.1016/j.cell.2011.10.002
Jiang Q, Ma R, Wang J et al (2015) LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC Genomics 16(3):1–11. https://doi.org/10.1186/1471-2164-16-S3-S2
Jiang C, Li Y, Zhao Z et al (2016) Identifying and functionally characterizing tissue-specific and ubiquitously expressed human lncRNAs. Oncotarget 7(6):7120–7133. https://doi.org/10.18632/oncotarget.6859
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821. https://doi.org/10.1126/science.1225829
Joaquina Delás M, Sabin LR, Dolzhenko E et al (2017) IncRNA requirements for mouse acute myeloid leukemia and normal differentiation. Elife. https://doi.org/10.7554/eLife.25607
Johnsson P, Lipovich L, Grandér D, Morris KV (2014) Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta - Gen Subj 1840(3):1063–1071. https://doi.org/10.1016/j.bbagen.2013.10.035
Kindgren P, Ard R, Ivanov M, Marquardt S (2018) Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation. Nat Commun 9(1):1–11. https://doi.org/10.1038/s41467-018-07010-6
Kosicki M, Tomberg K, Bradley A (2018) Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36(8):765–771. https://doi.org/10.1038/nbt.4192
Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, Pertea M (2019) Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol 20(1):1–13. https://doi.org/10.1186/s13059-019-1910-1
Kraft K, Geuer S, Will AJ et al (2015) Deletions, inversions, duplications: engineering of structural variants using CRISPR/Cas in mice. Cell Rep 10(5):833–839. https://doi.org/10.1016/j.celrep.2015.01.016
Kung JTY, Colognori D, Lee JT (2013) Long noncoding RNAs: past, present, and future. Genetics 193(3):651–669. https://doi.org/10.1534/genetics.112.146704
Kushawah G, Hernandez-Huertas L, Abugattas-Nuñez del Prado J et al (2020) CRISPR-Cas13d induces efficient mRNA knockdown in animal embryos. Dev Cell 54(6):805–817. https://doi.org/10.1016/j.devcel.2020.07.013
Labuhn M, Adams FF, Ng M et al (2018) Refined sgRNA efficacy prediction improves largeand small-scale CRISPR-Cas9 applications. Nucleic Acids Res 46(3):1375–1385. https://doi.org/10.1093/nar/gkx1268
Lagarde J, Uszczynska-Ratajczak B, Carbonell S et al (2017) High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat Genet 49(12):1731–1740. https://doi.org/10.1038/ng.3988
Lanzós A, Carlevaro-Fita J, Mularoni L et al (2017) Discovery of cancer driver long noncoding RNAs across 1112 tumour genomes: new candidates and distinguishing features. Sci Rep 7(1):1–16. https://doi.org/10.1038/srep41544
Lavalou P, Eckert H, Damy L et al (2019) Strategies for genetic inactivation of long noncoding RNAs in zebrafish. RNA 25(8):897–904. https://doi.org/10.1261/rna.069484.118
Leibowitz ML, Papathanasiou S, Doerfler PA et al (2021) Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat Genet 53(6):895–905. https://doi.org/10.1038/s41588-021-00838-7
Lekka E, Hall J (2018) Noncoding RNAs in disease. FEBS Lett 592(17):2884–2900. https://doi.org/10.1002/1873-3468.13182
Lennox KA, Behlke MA (2016) Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res 44(2):863–877. https://doi.org/10.1093/nar/gkv1206
Leucci E, Vendramin R, Spinazzi M et al (2016) Melanoma addiction to the long non-coding RNA SAMMSON. Nature 531(7595):518–522. https://doi.org/10.1038/nature17161
Lin N, Chang KY, Li Z et al (2014) An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 53(6):1005–1019. https://doi.org/10.1016/j.molcel.2014.01.021
Liu SJ, Horlbeck MA, Cho SW et al (2017) CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. https://doi.org/10.1126/science.aah7111
Liu Y, Cao Z, Wang Y et al (2018) Genome-wide screening for functional long noncoding RNAs in human cells by Cas9 targeting of splice sites. Nat Biotechnol 36(12):1203–1210. https://doi.org/10.1038/nbt.4283
Liu B, Luo Z, He J (2020) sgRNA-PSM: predict sgRNAs on-target activity based on position-specific mismatch. Mol Ther - Nucleic Acids 20:323–330. https://doi.org/10.1016/j.omtn.2020.01.029
Lord CJ, McDonald S, Swift S, Turner NC, Ashworth A (2008) A high-throughput RNA interference screen for DNA repair determinants of PARP inhibitor sensitivity. DNA Repair (amst) 7(12):2010–2019. https://doi.org/10.1016/j.dnarep.2008.08.014
Ma L, Cao J, Liu L et al (2019) Lncbook: a curated knowledgebase of human long non-coding rnas. Nucleic Acids Res 47(D1):D128–D134. https://doi.org/10.1093/nar/gky960
Maamar H, Cabili MN, Rinn J, Raj A (2013) linc-HOXA1 is a noncoding RNA that represses HoXa1 transcription in cis. Genes Dev 27(11):1260–1271. https://doi.org/10.1101/gad.217018.113
Mali P, Yang L, Esvelt KM et al (2013) RNA-guided human genome engineering via Cas9. Science 339(6121):823–826. https://doi.org/10.1126/science.1232033
Marchese FP, Raimondi I, Huarte M (2017) The multidimensional mechanisms of long noncoding RNA function. Genome Biol 18(1):1–13. https://doi.org/10.1186/s13059-017-1348-2
Mattioli K, Volders P-J, Gerhardinger C et al (2019) High-throughput functional analysis of lncRNA core promoters elucidates rules governing tissue specificity. Genome Res 29(3):344–355. https://doi.org/10.1101/GR.242222.118
Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F (2015) Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 518(7539):409–412. https://doi.org/10.1038/nature13975
Minotti L, Agnoletto C, Baldassari F, Corrà F, Volinia S (2018) SNPs and somatic mutation on long non-coding RNA: new frontier in the cancer studies? High-Throughput. https://doi.org/10.3390/HT7040034
Mizuno-Iijima S, Ayabe S, Kato K et al (2020) Efficient production of large deletion and gene fragment knock-in mice mediated by genome editing with Cas9-mouse Cdt1 in mouse zygotes. Methods 191:23–31. https://doi.org/10.1016/j.ymeth.2020.04.007
Mondal T, Subhash S, Vaid R et al (2015) MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA–DNA triplex structures. Nat Commun 6(1):1–17. https://doi.org/10.1038/ncomms8743
Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2(4):279–289. https://doi.org/10.1105/tpc.2.4.279
Necsulea A, Soumillon M, Warnefors M et al (2014) The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505(7485):635–640. https://doi.org/10.1038/nature12943
Palaz F, Kerem Kalkan A, Demir N, Tozluyurt A, Ozsoz M (2021) CRISPR-Cas13 system as a promising and versatile tool for cancer diagnosis, therapy, and research. ACS Synth Biol 10:1245–1267. https://doi.org/10.1021/acssynbio.1c00107
Palazzo AF, Lee ES (2015) Non-coding RNA: what is functional and what is junk? Front Genet 5:2. https://doi.org/10.3389/fgene.2015.00002
Pallarès-Albanell J, Zomeño-Abellán MT, Escaramís G et al (2019) a high-throughput screening identifies microRNA inhibitors that influence neuronal maintenance and/or response to oxidative stress. Mol Ther Nucleic Acids 17:374–387. https://doi.org/10.1016/j.omtn.2019.06.007
Perez AR, Pritykin Y, Vidigal JA et al (2017) GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol 35(4):347–349. https://doi.org/10.1038/nbt.3804
Perron U, Provero P, Molineris I (2017) In silico prediction of lncRNA function using tissue specific and evolutionary conserved expression. BMC Bioinformatics 18(5):14–16. https://doi.org/10.1186/s12859-017-1535-x
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33(3):290–295. https://doi.org/10.1038/nbt.3122
Pulido-Quetglas C, Aparicio-Prat E, Arnan C et al (2017) Scalable design of paired CRISPR guide RNAs for genomic deletion. PLOS Comput Biol 13(3):e1005341. https://doi.org/10.1371/journal.pcbi.1005341
Pyfrom SC, Luo H, Payton JE (2019) PLAIDOH: a novel method for functional prediction of long non-coding RNAs identifies cancer-specific LncRNA activities. BMC Genomics 20(1):1–24. https://doi.org/10.1186/s12864-019-5497-4
Qi LS, Larson MH, Gilbert LA et al (2013) Repurposing CRISPR as an RNA-γuided platform for sequence-specific control of gene expression. Cell 152(5):1173–1183. https://doi.org/10.1016/j.cell.2013.02.022
Radzisheuskaya A, Shlyueva D, Müller I, Helin K (2016) Optimizing sgRNA position markedly improves the efficiency of CRISPR/dCas9-mediated transcriptional repression. Nucleic Acids Res 44(18):e141. https://doi.org/10.1093/nar/gkw583
Ramilowski JA, Yip CW, Agrawal S et al (2020) Functional annotation of human long noncoding RNAs via molecular phenotyping. Genome Res 30(7):1060–1072. https://doi.org/10.1101/gr.254219.119
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8(11):2281–2308. https://doi.org/10.1038/nprot.2013.143
Rao DD, Vorhies JS, Senzer N, Nemunaitis J (2009) siRNA vs shRNA: similarities and differences. Adv Drug Deliv Rev 61(9):746–759. https://doi.org/10.1016/j.addr.2009.04.004
Roberts A, Pimentel H, Trapnell C, Pachter L (2011) Identification of novel transcripts in annotated genomes using RNA-seq. Bioinformatics 27(17):2325–2329. https://doi.org/10.1093/bioinformatics/btr355
Ruan X, Li P, Chen Y et al (2020) In vivo functional analysis of non-conserved human lncRNAs associated with cardiometabolic traits. Nat Commun 11(1):1–13. https://doi.org/10.1038/s41467-019-13688-z
Ruiz-Orera J, Albà MM (2019) Conserved regions in long non-coding RNAs contain abundant translation and protein–RNA interaction signatures. NAR Genomics Bioinform 1(1):e2–e2. https://doi.org/10.1093/nargab/lqz002
Sander JD, Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32(4):347–350. https://doi.org/10.1038/nbt.2842
Sanson KR, Hanna RE, Hegde M et al (2018) Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 9(1):1–15. https://doi.org/10.1038/s41467-018-07901-8
Sarropoulos I, Marin R, Cardoso-Moreira M, Kaessmann H (2019) Developmental dynamics of lncRNAs across mammalian organs and species. Nature 571(7766):510–514. https://doi.org/10.1038/s41586-019-1341-x
Schmitt AM, Chang HY (2017) Long noncoding RNAs: at the intersection of cancer and chromatin biology. Cold Spring Harb Perspect Med 7(7):1–16. https://doi.org/10.1101/cshperspect.a026492
Schmitz SU, Grote P, Herrmann BG (2016) Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci 73(13):2491–2509. https://doi.org/10.1007/s00018-016-2174-5
Seifuddin F, Singh K, Suresh A et al (2020) lncRNAKB, a knowledgebase of tissue-specific functional annotation and trait association of long noncoding RNA. Sci Data 7(1):1–16. https://doi.org/10.1038/s41597-020-00659-z
Seiler J, Breinig M, Caudron-Herger M, Polycarpou-Schwarz M, Boutros M, Diederichs S (2017) The lncRNA VELUCT strongly regulates viability of lung cancer cells despite its extremely low abundance. Nucleic Acids Res 45(9):5458–5469. https://doi.org/10.1093/nar/gkx076
Shalem O, Sanjana NE, Hartenian E et al (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343(6166):84–87. https://doi.org/10.1126/science.1247005
Shalem O, Sanjana NE, Zhang F (2015) High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16(5):299–311. https://doi.org/10.1038/nrg3899
Sims D, Mendes-Pereira AM, Frankum J et al (2011) High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome Biol 12(10):1–13. https://doi.org/10.1186/gb-2011-12-10-r104
Smith JR, Maguire S, Davis LA et al (2008) Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells 26(2):496–504. https://doi.org/10.1634/stemcells.2007-0039
Smith I, Greenside PG, Natoli T et al (2017) Evaluation of RNAi and CRISPR technologies by large-scale gene expression profiling in the connectivity map. PLoS Biol. https://doi.org/10.1371/journal.pbio.2003213
Soifer HS, Koch T, Lai J, Hansen B, Hoeg A, Oerum H, Stein CA (2012) Silencing of gene expression by gymnotic delivery of antisense oligonucleotides. Methods Mol Biol 815:333–346. https://doi.org/10.1007/978-1-61779-424-7_25
Sparber P, Filatova A, Khantemirova M, Skoblov M (2019) The role of long non-coding RNAs in the pathogenesis of hereditary diseases. BMC Med Genomics 12(2):63–78. https://doi.org/10.1186/s12920-019-0487-6
Stein CA, Hansen JB, Lai J et al (2010) Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res 38(1):e3. https://doi.org/10.1093/NAR/GKP841
Stemmer M, Thumberger T, Del Sol KM, Wittbrodt J, Mateo JL (2015) CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE. https://doi.org/10.1371/journal.pone.0124633
Stojic L, Niemczyk M, Orjalo A et al (2016) Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat Commun 7(1):1–14. https://doi.org/10.1038/ncomms10406
Stojic L, Lun ATL, Mascalchi P et al (2020) A high-content RNAi screen reveals multiple roles for long noncoding RNAs in cell division. Nat Commun 11(1):1–21. https://doi.org/10.1038/s41467-020-14978-7
Sudbery I, Enright AJ, Fraser AG, Dunham I (2010) Systematic analysis of off-target effects in an RNAi screen reveals microRNAs affecting sensitivity to TRAIL-induced apoptosis. BMC Genomics 11(1):1–12. https://doi.org/10.1186/1471-2164-11-175
Sun N, Petiwala S, Wang R et al (2019) Development of drug-inducible CRISPR-Cas9 systems for large-scale functional screening. BMC Genomics 20(1):1–15. https://doi.org/10.1186/s12864-019-5601-9
Sundaram A, Tengs T, Grimholt U (2017) Issues with RNA-seq analysis in non-model organisms: a salmonid example. Dev Comp Immunol 75:38–47. https://doi.org/10.1016/j.dci.2017.02.006
Thapar R (2018) Regulation of DNA double-strand break repair by non-coding RNAs. Molecules. https://doi.org/10.3390/molecules23112789
Tiessen I, Abildgaard MH, Lubas M et al (2019) A high-throughput screen identifies the long non-coding RNA DRAIC as a regulator of autophagy. Oncogene 38(26):5127–5141. https://doi.org/10.1038/s41388-019-0783-9
Trapnell C, Williams BA, Pertea G et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515. https://doi.org/10.1038/nbt.1621
Tsherniak A, Vazquez F, Montgomery PG et al (2017) Defining a cancer dependency map. Cell 170(3):564-576.e16. https://doi.org/10.1016/j.cell.2017.06.010
Turton N, Swan R, Mahenthiralingam T, Pitts D, Dykes IM (2019) The functions of long non-coding RNA during embryonic cardiovascular development and its potential for diagnosis and treatment of congenital heart disease. J Cardiovasc Dev Dis 6(2):21. https://doi.org/10.3390/jcdd6020021
Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147(7):1537–1550. https://doi.org/10.1016/j.cell.2011.11.055
Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigó R, Johnson R (2018) Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet 19(9):535–548. https://doi.org/10.1038/s41576-018-0017-y
Vancura A, Lanzós A, Bosch-Guiteras N et al (2021) Cancer LncRNA Census 2 (CLC2): an enhanced resource reveals clinical features of cancer lncRNAs. NAR Cancer. https://doi.org/10.1093/narcan/zcab013
Vendramin R, Verheyden Y, Ishikawa H et al (2018) SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nat Struct Mol Biol 25(11):1035–1046. https://doi.org/10.1038/s41594-018-0143-4
Vidigal JA, Ventura A (2015) Rapid and efficient one-step generation of paired gRNA CRISPR-Cas9 libraries. Nat Commun. https://doi.org/10.1038/ncomms9083
Wang T, Wei JJ, Sabatini DM, Lander ES (2014) Genetic screens in human cells using the CRISPR-Cas9 system. Science 343(6166):80–84. https://doi.org/10.1126/science.1246981
Wang Y, Juan L, Peng J, Zang T, Wang Y (2019) LncDisAP: a computation model for LncRNA-disease association prediction based on multiple biological datasets. BMC Bioinform. https://doi.org/10.1186/s12859-019-3081-1
Washietl S, Kellis M, Garber M (2014) Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res 24(4):616–628. https://doi.org/10.1101/gr.165035.113
Wen K, Yang L, Xiong T et al (2016) Critical roles of long noncoding RNAs in Drosophila spermatogenesis. Genome Res 26(9):1233–1244. https://doi.org/10.1101/gr.199547.115
Whitehurst AW, Bodemann BO, Cardenas J et al (2007) Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 446(7137):815–819. https://doi.org/10.1038/nature05697
Wong N, Liu W, Wang X (2015) WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol 16(1):1–8. https://doi.org/10.1186/s13059-015-0784-0
Wu X, Kriz AJ, Sharp PA (2014) Target specificity of the CRISPR-Cas9 system. Quant Biol 2(2):59. https://doi.org/10.1007/S40484-014-0030-X
Xiong H, Veedu RN, Diermeier SD (2021) Recent advances in oligonucleotide therapeutics in oncology. Int J Mol Sci. https://doi.org/10.3390/ijms22073295
Xu H, Xiao T, Chen CH et al (2015) Sequence determinants of improved CRISPR sgRNA design. Genome Res 25(8):1147–1157. https://doi.org/10.1101/gr.191452.115
Xu D, Cai Y, Tang L et al (2020) A CRISPR/Cas13-based approach demonstrates biological relevance of vlinc class of long non-coding RNAs in anticancer drug response. Sci Rep 10(1):1–13. https://doi.org/10.1038/s41598-020-58104-5
Yang LZ, Wang Y, Li SQ et al (2019) Dynamic imaging of RNA in living cells by CRISPR-Cas13 systems. Mol Cell 76(6):981-997.e7. https://doi.org/10.1016/j.molcel.2019.10.024
Yoshida T, Naito Y, Yasuhara H et al (2019) Evaluation of off-target effects of gapmer antisense oligonucleotides using human cells. Genes Cells 24(12):827–835. https://doi.org/10.1111/gtc.12730
Zeng Y, Cullen BR (2002) RNA interference in human cells is restricted to the cytoplasm. RNA 8(7):855–860. https://doi.org/10.1017/S1355838202020071
Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH (2015) Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids 4(11):e264. https://doi.org/10.1038/mtna.2015.37
Zhao H, Shi J, Zhang Y et al (2020) LncTarD: a manually-curated database of experimentally-supported functional lncRNA-target regulations in human diseases. Nucleic Acids Res 48(D1):D118–D126. https://doi.org/10.1093/nar/gkz985
Zheng Q, Cai X, Tan MH et al (2014) Precise gene deletion and replacement using the CRISPR/Cas9 system in human cells. Biotechniques 57(3):115–124. https://doi.org/10.2144/000114196
Zhu S, Li W, Liu J et al (2016) Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol 34(12):1279–1286. https://doi.org/10.1038/nbt.3715
Zhu S, Cao Z, Liu Z et al (2019) Guide RNAs with embedded barcodes boost CRISPR-pooled screens. Genome Biol 20(1):1–12. https://doi.org/10.1186/s13059-019-1628-0
Acknowledgements
We acknowledge administrative and logistical support from Basak Ginsbourger, Rahel Tschudi, Marla Rittiner, Beatrice Stalder, Willy Hofstetter and Patrick Furer (DBMR, University of Bern). We thank Panagiotis Chouvardas and other members of GOLD Lab for insightful feedback and discussions. Work in the GOLD Lab is funded by the Swiss National Science Foundation through the National Center of Competence in Research (NCCR) “RNA & Disease,” by the Medical Faculty of the University and University Hospital of Bern, by the Helmut Horten Stiftung, Swiss Cancer Research Foundation (4534-08-2018), and Science Foundation Ireland through Future Research Leaders award 18/FRL/6194.
Funding
Open Access funding provided by Universität Bern.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pulido-Quetglas, C., Johnson, R. Designing libraries for pooled CRISPR functional screens of long noncoding RNAs. Mamm Genome 33, 312–327 (2022). https://doi.org/10.1007/s00335-021-09918-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-021-09918-9