Abstract
Ferns are a large and evolutionarily critical group of vascular land plants for which quite limited mitochondrial gene content and genome organization data are, currently, available. This study reports that the gene for the ribosomal protein S3 (rps3) is preserved and physically clustered to an upstream rps19 and a downstream overlapping rpl16 locus in the mitochondrial DNA of the true fern Adiantum capillus-veneris L. Sequence analysis also revealed that the rps3 gene is interrupted by two cis-splicing group II introns, like the counterpart in lycopod and gymnosperm representatives. A preliminary polymerase chain reaction (PCR) survey confirmed a scattered distribution pattern of both the rps3 introns also in other fern lineages. Northern blot and reverse transcription (RT)–PCR analyses demonstrated that the three ribosomal protein genes are co-transcribed as a polycistronic mRNA and modified by RNA editing. Particularly, the U-to-C type editing amends numerous genomic stop codons in the A. capillus-veneris rps19, rps3 and rpl16 sequences, thus, assuring the synthesis of complete and functional polypeptides. Collectively, the findings from this study further expand our knowledge of the mitochondrial rps3 architecture and evolution, also, bridging the significant molecular data gaps across the so far underrepresented ferns and all land plants.



Similar content being viewed by others
References
Adams KL, Palmer JD (2003) Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol 29:380–395
Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD (2010) Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol 27:1436–1448
Alverson AJ, Rice DW, Dickinson S, Barry K, Palmer JD (2011) Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell 23:2499–2513
Bégu D, Araya A (2009) The horsetail Equisetum arvense mitochondria share two group I introns with the liverwort Marchantia, acquired a novel group II intron but lost intron-encoded ORFs. Curr Genet 55:69–79
Bégu D, Castandet B, Araya A (2011) RNA editing restores critical domains of a group I intron in fern mitochondria. Curr Genet 57:317–325
Bernt M, Braband A, Schierwater B, Stadler PF (2013) Genetic aspects of mitochondrial genome evolution. Mol Phylogenet Evol 69:328–338
Binder S, Hölzle A, Jonietz C (2010) RNA processing and RNA stability in plant mitochondria. In: Kempken F (ed) plant mitochondria. Springer Science and Business Media, New York, pp 107–130
Bonen L (2008) Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8:26–34
Bonen L (2012) Evolution of mitochondrial introns in plants and photosynthetic microbes. Adv Bot Res 63:155–186. doi:10.1016/B978-0-12-394279-1.00007-7
Christenhusz MJM, Chase MW (2014) Trends and concepts of fern classification. Ann Bot 113:571–594. doi:10.1093/aob/mct299
Darracq A, Varré JS, Maréchal-Drouard L, Courseaux A, Castric V, Saumitou-Laprade P, Oztas S, Lenoble P, Vacherie B, Barbe V, Touzet P (2011) Structural and content diversity of mitochondrial genome in beet: a comparative genomic analysis. Genome Biol Evol 3:723–736
Fedorova O, Zingler N (2007) Group II introns: structure, folding and splicing mechanism. Biol Chem 388:665–678
Grewe F, Viehoever P, Weisshaar B, Knoop V (2009) A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res 37:5093–5104
Grewe F, Herres S, Viehöver P, Polsakiewicz M, Weisshaar B, Knoop V (2011) A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res 39:2890–2902
Hecht J, Grewe F, Knoop V (2011) Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol 3:344–358
Hegedusova E, Brejova B, Tomaska L, Sipiczki M, Nosek J (2014) Mitochondrial genome of the basidiomycetous yeast Jaminaea angkorensis. Curr Genet 60:49–59
Himmelstrand K, Olson A, Brandström Durling M, Karlsson M, Stenlid J (2014) Intronic and plasmid–derived regions contribute to the large mitochondrial genome sizes of Agaricomycetes. Curr Genet 60:303–313
Hunt MD, Newton KJ (1991) The NCS3 mutation: genetic evidence for the expression of ribosomal protein genes in Zea mays mitochondria. EMBO J 10:1045–1052
Knoop V (2004) The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet 46:123–139
Knoop V (2011) When you can’t trust the DNA: RNA editing changes transcript sequences. Cell Mol Life Sci 68:567–586
Kubo N, Ozawa K, Hino T, Kadowaki K (1996) A ribosomal protein L2 gene is transcribed, spliced, and edited at one site in rice mitochondria. Plant Mol Biol 31:853–862
Laroche J, Bousquet J (1999) Evolution of the mitochondrial rps3 intron in perennial and annual angiosperms and homology to nad5 intron 1. Mol Biol Evol 16:441–452
Liu Y, Xue J-Y, Wang B, Li L, Qiu Y-L (2011) The mitochondrial genomes of the early land plants Treubia lacunose and Anomodon rugelii: dynamic and conservative evolution. PLoS One 6:e25836
Liu Y, Wang B, Cui P, Li L, Xue JY, Yu J, Qiu YL (2012) The mitochondrial genome of the lycophyte Huperzia squarrosa: the most archaic form in vascular plants. PLoS One 7:e35168
Martínez-Abarca F, Toro N (2000) Group II introns in the bacterial world. Mol Microbiol 38:917–926
Michel F, Umesono K, Ozeki H (1989) Comparative and functional anatomy of group II catalytic introns. Gene 82:5–30
Nakazono M, Itadani H, Wakasugi T, Tsutsumi N, Sugiura M, Hirai A (1995) The rps3-rpl16-nad3-rps12 gene cluster in rice mitochondrial DNA is transcribed from alternative promoters. Curr Genet 27:184–189
Oda K, Yamoto K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Kanegae T, Ogura Y, Kohchi T, Ohyama K (1992) Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. J Mol Biol 223:1–7
Perrotta G, Grienenberger JM, Gualberto JM (2002) Plant mitochondrial rps2 genes code for proteins with a C-terminal extension that is processed. Plant Mol Biol 50:523–533
Pryer KM, Schuettpelz E, Wolf PG, Schneider H, Smith AR, Cranfill R (2004) Phylogeny and evolution of ferns (Monilophytes) with a focus on the early leptosporangiate divergences. Am J Bot 91:1582–1598
Ran JH, Gao H, Wang XQ (2010) Fast evolution of the retroprocessed mitochondrial rps3 gene in Conifer II and further evidence for the phylogeny of gymnosperms. Mol Phylogenet Evol 54:136–149
Regina TMR, Quagliariello C (2010) Lineage-specific group II intron gains and losses of the mitochondrial rps3 gene in gymnosperms. Plant Physiol Biochem 48:646–654
Regina TMR, Picardi E, Lopez L, Pesole G, Quagliariello C (2005) A novel additional group II intron distinguishes the mitochondrial rps3 gene in gymnosperms. J Mol Evol 60:196–206
Rice DW, Alverson AJ, Richardson AO, Young GJ, Sanchez-Puerta MV, Munzinger J, Barry K, Boore JL, Zhang Y, dePamphilis CW, Knox EB, Palmer JD (2013) Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 342:1468–1473
Richardson AO, Palmer JD (2007) Horizontal gene transfer in plants. J Exp Bot 58:1–9
Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD (2013) The “fossilized” mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol 11:29
Robart AR, Zimmerly S (2005) Group II intron retroelements: function and diversity. Cytogenet Genome Res 110:589–597
Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG (2014) From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol 14:23
Schuettpelz E, Pryer KM (2009) Evidence for a Cenozoic radiation of ferns in an angiosperm-dominated canopy. Proc Natl Acad Sci USA 106:11200–11205
Schuster W, Brennicke A (1991) RNA editing makes mistakes in plant mitochondria: editing loses sense in transcripts of a rps19 pseudogene and in creating stop codons in coxI and rps3 mRNAs of Oenothera. Nucleic Acids Res 19:6923–6928
Sloan DB, Alverson AJ, Štorchová H, Palmer JD, Taylor DR (2010) Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia. BMC Evol Biol 10:274
Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, Palmer JD, Taylor DR (2012a) Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol 10:e1001241
Sloan DB, Müller K, McCauley DE, Taylor DR, Storchová H (2012b) Intraspecific variation in mitochondrial genome sequence, structure, and gene content in Silene vulgaris, an angiosperm with pervasive cytoplasmic male sterility. New Phytol 196:1228–1239. doi:10.1111/j.1469-8137.2012.04340.x
Smith AR, Pryer KM, Schuettpelz E, Korall P, Schneider H, Wolf PG (2006) A classification for extant ferns. Taxon 55:705–731
Takemura M, Oda K, Yamato K, Otha E, Nakamura Y, Nozato N, Akashi K, Ohyama K (1992) Gene clusters for ribosomal proteins in the mitochondrial genome of a liverwort, Marchantia polymorpha. Nucleic Acids Res 20:3199–3205
Takenaka M (2014) How complex are the editosomes in plant organelles? Mol Plant 7:582–585
Takenaka M, Zehrmann A, Verbitskiy D, Härtel B, Brennicke A (2013) RNA Editing in Plants and Its Evolution. Annu Rev Genet 47:335–352
Tanaka Y, Tsuda M, Yasumoto K, Terachi T, Yamagishi H (2014) The complete mitochondrial genome sequence of Brassica oleracea and analysis of coexisting mitotypes. Curr Genet 60:277–284
Terasawa K, Odahara M, Kabeya Y, Kikugawa T, Sekine Y, Fujiwara M, Sato N (2007) The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol Biol Evol 24:699–709
Turmel M, Otis C, Lemieux C (2003) The mitochondrial genome of Chara vulgaris: insights into the mitochondrial DNA architecture of the last common ancestor of green algae and land plants. Plant Cell 15:1888–1903
Turmel M, Otis C, Lemieux C (2013) Tracing the evolution of streptophyte algae and their mitochondrial genome. Genome Biol Evol 5:1817–1835
Wolf PG, Rowe CA, Sinclair RB, Hasebe M (2003) Complete nucleotide sequence of the chloroplast genome from a leptosporangiate fern, Adiantum capillus-veneris L. DNA 10:59–65
Wolf PG, Rowe CA, Hasebe M (2004) High levels of RNA editing in a vascular plant chloroplast genome: analysis of transcripts from the fern Adiantum capillus-veneris. Gene 339:89–97
Acknowledgments
We thank the Orto Botanico at the Università degli Studi di Napoli and Calabria for kindly made available green leaf tissue from the analyzed plants. We also acknowledge the cooperation with Laszlo Csiba at the Royal Botanic Gardens at Kew (U.K.) for providing some of the DNAs used in this study. Work in the authors’ laboratory was supported by grants from the Università della Calabria and the Italian Ministero dell’Istruzione, Università e Ricerca (M.I.U.R.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by M. Kupiec.
This article is dedicated to the memory of Prof. Carla Quagliariello (1949–2012).
Electronic supplementary material
Below is the link to the electronic supplementary material.
294_2015_512_MOESM1_ESM.docx
Supplementary material 1 (DOCX 24 kb) Supplemental S1 List of oligonucleotides used as primers for PCR and RT-PCR amplification and sequencing
294_2015_512_MOESM2_ESM.tif
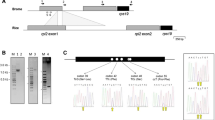
Supplementary material 2 (TIFF 104 kb) Supplemental S2 Nucleotide and derived amino acid sequences of the mitochondrial rps3 gene from A. capillus-veneris. The amino acid sequence is indicated using a single letter abbreviation above the nucleotide sequence. Nucleotides altered by RNA editing, corresponding to C-to-U and U-to-C changes at the RNA level, are indicated in lower-case letters and underlined, whereas the resulting amino acid substitutions (unedited > edited amino acid residues) are given in bold. Vertical arrows indicate the insertion site of the rps3 introns. The position and the orientation of some primers (P1 - P4) used for PCR and RT-PCR amplification and sequencing reactions (see Fig. 1c) are indicated in italic letters and in bold. The A. capillus-veneris nucleotides and amino acids are numbered on the right
294_2015_512_MOESM3_ESM.tif
Supplementary material 3 (TIFF 132 kb) Supplemental S3 RNA secondary structure model of the mitochondrial rps3i1 (a) and rps3i2 (b) introns from A. capillus-veneris. According to Michel et al. (1989), both the two rps3 introns are organized into six well-defined domains (dI–dVI) originating from a central wheel. Exon and intron binding sites (EBS and IBS), the branching point adenosine (marked by an asterisk in domain VI) and the γ-γ′ interactions are indicated. Loops are not drawn to scale and numbers inside the loops indicate their size
Rights and permissions
About this article
Cite this article
Bonavita, S., Regina, T.M.R. The evolutionary conservation of rps3 introns and rps19-rps3-rpl16 gene cluster in Adiantum capillus-veneris mitochondria. Curr Genet 62, 173–184 (2016). https://doi.org/10.1007/s00294-015-0512-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-015-0512-z