Abstract
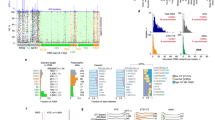
5S ribosomal RNA (rRNA) genes are known to move in and out of various tandemly repeated eukaryotic gene families during evolution. Here, we investigated the organization of 5S rRNA genes linked to the ribosomal DNA (rDNA) units in 147 fungal species using available sequence and genome databanks. Whereas some fungal species have no 5S rRNA genes in their rDNA units, others have one or two 5S rRNA copies linked on the same or the opposite strand. There were at least 13 independent changes during the evolution of fungal species. These include two 5S rRNA genes loss, five 5S rRNA genes inversions and six 5S rRNA genes insertions (including duplications). The lower frequency of 5S rRNA genes loss might be due to the fact that these events are more likely to affect fitness. The maximum time required for 5S rRNA gene organization to change between related species was estimated to be 7.5 millions years based on the sequences of the elongation factor alpha genes of Candida glabrata and Saccharomyces mikatae. This time is much longer than the homogenization time predicted from theoretical and experimental studies and likely reflects the lack of closely related species or strains in our data set.



Similar content being viewed by others
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Arnheim N (1983) Concerted evolution of multigene families. In: Nei M, Koehn R (eds) Evolution of genes and proteins. Sinauer, Sunderland, pp 38–61
Babasaki K, Neda H, Murata H (2007) megB1, a novel macroevolutionary genomic marker of the fungal phylum Basidiomycota. Biosci Biotechnol Biochem 71:1927–1939
Bedard JE, Schurko AM, de Cock AW, Klassen GR (2006) Diversity and evolution of 5S rRNA gene family organization in Pythium. Mycol Res 110:86–95
Begerow D, Stoll M, Bauer R (2006) A phylogenetic hypothesis of Ustilaginomycotina based on multiple gene analyses and morphological data. Mycologia 98:909–916
Belkhiri A, Buchko J, Klassen GR (1992) The 5S ribosomal RNA gene in Pythium species: two different genomic locations. Mol Biol Evol 9:1089–1102
Berbee ML, Taylor JW (2001) Fungal molecular evolution: gene trees and geologic time. In: McLaughlin DJ, McLaughlin EG, Lemke PA (eds) The Mycota VIIB systematics and evolution. Springer, Berlin, pp 229–245
Cai J, Roberts IN, Collins MD (1996) Phylogenetic relationships among members of the ascomycetous yeast genera Brettanomyces Debaryomyces, Dekkera, and Kluyveromyces deduced by small-subunit rRNA gene sequences. Int J Syst Bacteriol 46:542–549
Diezmann S, Cox CJ, Schönian G, Vilgalys RJ, Mitchell TG (2004) Phylogeny and evolution of medical species of Candida and related taxa: a multigenic analysis. J Clin Microbiol 42:5624–5635
Dover GA (1982) Molecular drive: a cohesive mode of species evolution. Nature 299:111–117
Drouin G, Moniz de Sá M (1995) The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. Mol Biol Evol 12:481–493
Drouin G (2000) Expressed retrotransposed 5S rRNA genes in the mouse and rat genomes. Genome 43:213–215
Easteal S, Collet CC, Betty DJ (1995) The mammalian molecular clock. Springer & Landes, Austin
Fitzpatrick DA, Loque ME, Stajich JE, Butler G (2006) A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol 6:99
Ganley AR, Kobayashi T (2007) Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res 17:184–191
Gaudet G, Julien J, Lafay JF, Brygoo Y (1989) Phylogeny of some Fusarium species, as determined by large-subunit rRNA sequence comparison. Mol Biol Evol 6:227–242
Gerbi S (1985) Evolution of ribosomal DNA. In: MacIntyre RJ (ed) Molecular evolutionary genetics. Plenum, New York, pp 419–517
Glazko GV, Koonin EV, Rogozin IB (2005) Molecular dating: Ape bones agree with chicken entrails. Trends Genet 21:89–92
Graur D, Li W-H (2000) Fundamentals of molecular evolution, 2nd edn. Sinauer Associates, Sunderland
Graur D, Martin W (2004) Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision. Trends Genet 20:242–247
Guerreiro P, Neves A, Rodrigues-Pousada C (1993) Clusters of 5S rRNAs in the intergenic region of ubiquitin genes in Tetrahymena pyriformis. Biochim Biophys Acta 1216:137–139
Hinkle G, Wetterer JK, Schultz TR, Sogin ML (1994) Phylogeny of the Attine ant fungi based on analysis of the small subunit ribosomal RNA gene sequences. Science 266:1695–1697
James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüssler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lücking R, Büdel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R (2006) Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443:818–822
Kasuga T, White TJ, Taylor JW (2002) Estimation of nucleotide substitution rates in Eurotiomycete fungi. Mol Biol Evol 19:2318–2324
Kurtzman CP (2000) Four new yeasts in the Pichia anomala clade. Int J Syst Evol Microbiol 50:395–404
Kurtzman CP, Robnett CJ (2003) Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res 3:417–432
Kyei-Poku G, Gauthier D, Van Frankenhuyzen K (2008) Molecular data and phylogeny of Nosema infecting Lepidopteran forest defoliators in the genera Choristoneura and Malacosoma. J Eukaryot Microbiol 55:51–58
Larrivée M, Wellinger RJ (2006) Telomerase- and capping-independent yeast survivors with alternate telomere states. Nat Cell Biol 8:741–747
Lévesque CA, de Cock AW (2004) Molecular phylogeny and taxonomy of the genus Pythium. Mycol Res 108:1363–1383
Li WH (1993) Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol 36:96–99
Lutzoni F, Kauff F, Cox CJ, McLaughlin D, Celio G, Dentinger B, Padamsee M, Hibbett D, James TY, Baloch E, Grube M, Reeb V, Hofstetter V, Schoch C, Arnold AE, Miadlikowska J, Spatafora J, Johnson D, Hambleton S, Crockett M, Shoemaker R, Sung G-H, Lücking R, Lumbsch T, O’Donnell K, Binder M, Diederich P, Ertz D, Gueidan C, Hansen K, Harris RC, Hosaka K, Matheny B, Nishida H, Pfister D, Rogers J, Rossman A, Schmitt I, Sipman H, Stone J, Sugiyama J, Yahr R, Vilgalys R (2004) Assembling the fungal tree of life: progress, classification and evolution of subcellular traits. Am J Bot 91:1446–1480
Manchado M, Zuasti E, Cross I, Merlo A, Infante C, Rebordinos L (2006) Molecular characterization and chromosomal mapping of the 5S rRNA gene in Solea senegalensis: a new linkage to the U1, U2, and U5 small nuclear RNA genes. Genome 49:79–86
Matheny PB, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson RH, Hughes KW, Hofstetter V, Ammirati JF, Schoch CL, Langer E, Langer G, McLaughlin DJ, Wilson AW, Frøslev T, Ge ZW, Kerrigan RW, Slot JC, Yang ZL, Baroni TJ, Fischer M, Hosaka K, Matsuura K, Seidl MT, Vauras J, Hibbett DS (2007) Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol Phylogenet Evol 43:430–451
Moncalvo JM, Lutzoni F, Rehner S, Johnson J, Vilgalys R (1996) Molecular phylogeny of the Agaricales based on 25S rDNA sequences. Department of Botany, Duke University. Presented as a poster at the Asilomar Fungal Genetics Conference (March 1996). http://www.biology.duke.edu/fungi/mycolab/agarical.htm
Nachman MW, Crowell SL (2000) Estimate of the mutation rate per nucleotide in humans. Genetics 156:297–304
Nazar RN (1980) A 5.8S rRNA-like sequence in prokaryotic 23S rRNA. FEBS Lett 119:212–214
Ochman H, Wilson AC (1987) Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol 26:74–86
Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Pamilo P, Bianchi NO (1993) Evolution of the Zfx and Zfy, genes: rates and interdependence between the genes. Mol Biol Evol 10:271–281
Pelliccia F, Barzotti R, Bucciarelli E, Rocchi A (2001) 5S ribosomal and U1 small nuclear RNA genes: a new linkage type in the genome of a crustacean that has three different tandemly repeated units containing 5S ribosomal DNA sequences. Genome 44:331–335
Rubin GM, Sulston JE (1973) Physical linkage of the 5 S cistrons to the 18 S and 28 S ribosomal RNA cistrons in Saccharomyces cerevisiae. J Mol Biol 79:521–530
Smith GP (1974) Unequal crossover and the evolution of multigene families. Cold Spring Harbor Symp Quant Biol 38:507–513
Souciet J, Aigle M, Artiguenave F, Blandin G, Bolotin-Fukuhara M, Bon E, Brottier P, Casaregola S, de Montigny J, Dujon B, Durrens P, Gaillardin C, Lépingle A, Llorente B, Malpertuy A, Neuvéglise C, Ozier-Kalogéropoulos O, Potier S, Saurin W, Tekaia F, Toffano-Nioche C, Wésolowski-Louvel M, Wincker P, Weissenbach J (2000) Genomic exploration of the hemiascomycetous yeasts: 1. A set of yeast species for molecular evolution studies. FEBS Lett 487:3–12
Szostak JW, Wu R (1980) Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature 284:426–430
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thon MR, Royse DJ (1999) Partial β-tubulin gene sequences for evolutionary studies in the Basidiomycotina. Mycologia 91:468–474
Tsui CKM, Daniel HM, Robert V, Meyer W (2008) Re-examining the phylogeny of clinically relevant Candida species and allied genera based on multigene analyses. FEMS Yeast Res 8:651–659
Voigt K, Cigelnik E, O’Donnell K (1999) Phylogeny and PCR identification of clinically important zygomycetes based on nuclear ribosomal-DNA sequence data. J Clin Microbiol 37:3957–3964
Wolfe KH, Sharp PM, Li WH (1989) Rates of synonymous substitutions in plant nuclear genes. J Mol Evol 29:208–211
Zuckerkandl E, Pauling L (1965) Molecules as documents of evolutionary history. J Theor Biol 8:357–366
Acknowledgments
We thank Mary Berbee (Botany Department, University of British Columbia) for her advice and comments on fungal divergence times. We thank the two anonymous reviewers for their thoughtful and constructive comments on a previous version of this manuscript. This work was supported by a Discovery Grant from the National Science and Engineering Research Council of Canada to G. D.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Gaillardin.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bergeron, J., Drouin, G. The evolution of 5S ribosomal RNA genes linked to the rDNA units of fungal species. Curr Genet 54, 123–131 (2008). https://doi.org/10.1007/s00294-008-0201-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-008-0201-2