Abstract
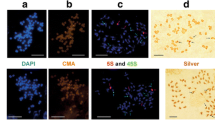
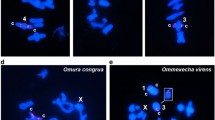
The 5S ribosomal DNA (rDNA) sequences are subject of dynamic evolution at chromosomal and molecular levels, evolving through concerted and/or birth-and-death fashion. Among grasshoppers, the chromosomal location for this sequence was established for some species, but little molecular information was obtained to infer evolutionary patterns. Here, we integrated data from chromosomal and nucleotide sequence analysis for 5S rDNA in two Abracris species aiming to identify evolutionary dynamics. For both species, two arrays were identified, a larger sequence (named type-I) that consisted of the entire 5S rDNA gene plus NTS (non-transcribed spacer) and a smaller (named type-II) with truncated 5S rDNA gene plus short NTS that was considered a pseudogene. For type-I sequences, the gene corresponding region contained the internal control region and poly-T motif and the NTS presented partial transposable elements. Between the species, nucleotide differences for type-I were noticed, while type-II was identical, suggesting pseudogenization in a common ancestor. At chromosomal point to view, the type-II was placed in one bivalent, while type-I occurred in multiple copies in distinct chromosomes. In Abracris, the evolution of 5S rDNA was apparently influenced by the chromosomal distribution of clusters (single or multiple location), resulting in a mixed mechanism integrating concerted and birth-and-death evolution depending on the unit.


Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Anjos A, Loreto V, de Souza MJ (2013) Chromosome mapping of ribosomal genes and histone H4 in the genus Radacridium (Romaleidae). Genet Mol Biol 36:336–340
Anjos A, Ruiz-Ruano FJ, Camacho JPM, Loreto V, Cabrero J, de Souza MJ, Cabral-de-Mello DC (2015) U1 snDNA clusters in grasshoppers: chromosomal dynamics and genomic organization. Heredity 114:207–219
Bueno D, Palacios-Gimenez OM, Cabral-de-Mello DC (2013) Chromosomal mapping of repetitive DNAs in Abracris flavolineata reveal possible ancestry for the B chromosome and surprisingly H3 histone spreading. PLoS ONE 8:e66532
Cabral-de-Mello DC, Moura RC, Martins C (2010) Chromosomal mapping of repetitive DNAs in the beetle Dichotomius geminatus provides the first evidence for an association of 5S rRNA and histone H3 genes in insects, and repetitive DNA similarity between the B chromosome and A complement. Heredity 104:393–400
Cabral-de-Mello DC, Cabrero J, López-León MD, Camacho JPM (2011a) Evolutionary dynamics of 5S rDNA location in acridid grasshoppers and its relationship with H3 histone gene and 45S rDNA location. Genetica 139:921–931
Cabral-de-Mello DC, Martins C, Souza MJ, Moura RC (2011b) Cytogenetic mapping of 5S and 18S rRNAs and H3 histone genes in 4 ancient Proscopiidae grasshopper species: contribution to understanding the evolutionary dynamics of multigene families. Cytogenet Genome Res 132:89–93
Castillo ER, Taffarel A, Marti DA (2011) Una técnica alternativa para el cariotipado mitótico en saltamontes: bandeo C y Fluorescente en Adimantus ornatissimus (Orthoptera: Acrididae). Rev Cienc Tecnol 16:30–34
Cross I, Rebordinos L (2005) 5S rDNA and U2 snRNA are linked in the genome of Crassostrea angulate and Crassostrea gigas oysters: does the (CT) n ·(GA) n microsatellite stabilize this novel linkage of large tandem arrays? Genome 48:1116–1119
da Silva M, Matoso DA, Vicari MR, de Almeida MC, Margarido VP, Artoni RF (2011) Physical mapping of 5S rDNA in two species of knifefishes: Gymnotus pantanal and Gymnotus paraguensis (Gymnotiformes). Cytogenet Genome Res 134:303–307
Drouin G, Moniz de Sá M (1995) The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. Mol Biol Evol 12:481–493
Drummond AJ, Ashton B, Cheung M, Heled J, Kearse M, Moir R et al (2009) Geneious v4.8.5. http://www.geneious.com
Haper ME, Price J, Korn LJ (1983) Chromosomal mapping of Xenopus 5S genes: somatic-type versus oocyte-type. Nucl Acids Res 11:2313–2323
Jensen LR, Frederiksen S (2000) The 5S rRNA genes in Macaca fascicularis are organized in two large tandem repeats. Biochim Biophys Acta 1492:537–542
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Little RD, Braaten DC (1989) Genomic organization of human 5S rDNA and sequence of one tandem repeat. Genomics 4:376–383
Long EO, Dawid IB (1980) Repeated genes in eukaryotes. Ann Rev Biochem 49:727–764
Martins C, Galetti PM Jr (2001) Organization of 5S rDNA in species of the fish Leporinus: two different genomic locations are characterized by distinct nontranscribed spacers. Genome 44:903–910
Martins C, Galleti PM Jr (2001) Two 5S rDNA arrays in neotropical fish species: is it a general rule for fishes? Genetica 111:439–446
Martins C, Wasko AP, Oliveira C, Porto-Foresti F, Parise-Maltempi PP, Wright JM, Foresti F (2002) Dynamics of 5S rDNA in the tilapia (Oreochromis niloticus) genome: repeat units, inverted sequences, pseudogenes and chromosome loci. Cytogenet Genome Res 98:78–85
Merlo MA, Cross I, Palazón JL, Úbeda-Manzanaro M, Sarasquete C, Rebordinos L (2012a) Evidence for 5S rDNA horizontal transfer in the toadfish Halobatrachus didactylus (Schneider, 1801) based on the analysis of three multigene families. BMC Evol Biol 12:201
Merlo MA, Pacchiarini T, Portela-Bens S, Cross I, Manchado M, Rebordinos L (2012b) Genetic characterization of Plectorhinchus mediterraneus yields important clues about genome organization and evolution of multigene families. BMC Genet 13:33
Merlo MA, Cross I, Manchado M, Cárdenas S, Rebordinos L (2013) The 5S rDNA high dynamism in Diplodus sargus is a transposon-mediated mechanism. Comparison with other multigene families and Sparidae species. J Mol Evol 76:83–97
Nederby-Nielsen J, Hallenberg C, Frederiksen S, Sorensen PD, Lomholt B (1993) Transcription of human 5S rRNA genes is influenced by an upstream DNA sequence. Nucleic Acids Res 21:3631–3636
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Ann Rev Genetics 39:121–152
Neto MSR, de Souza MJ, Loreto V (2013) Chromosomal evolution of rDNA and H3 histone genes in representative Romaleidae grasshoppers from northeast Brazil. Mol Cytogenetics 6:41
Palacios-Gimenez OM, Castillo ER, Martí DA, Cabral-de-Mello DC (2013) Tracking the evolution of sex chromosome systems in Melanoplinae grasshoppers through chromosomal mapping of repetitive DNA sequences. BMC Evol Biol 13:167
Perina A, Seoane D, González-Tizón AM, Rodríguez-Fariña F, Martínez-Lage A (2011) Molecular organization and phylogenetic analysis of 5S rDNA in crustaceans of the genus Pollicipes reveal birth-and-death evolution and strong purifying selection. BMC Evol Biol 11:304
Pinhal D, Yoshimura TS, Araki CS, Martins C (2011) The 5S rDNA family evolves through concerted and birth-and-death evolution in fish genomes: an example from freshwater stingrays. BMC Evol Biol 11:151
Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83:2934–2938
Rebordinos L, Cross I, Merlo A (2013) High evolutionary dynamism in 5S rDNA of fish: State of the Art. Cytogenet Genome Res 141:103–113
Rodrigues DS, Rivera M, Lourenço LB (2012) Molecular organization and chromosomal localization of 5S rDNA in Amazonian Engystomops (Anura, Leiuperidae). BMC Genet 13:17
Rooney AP, Ward TJ (2005) Evolution of a large ribosomal RNA multigene family in filamentous fungi: birth and death of a concerted evolution paradigm. Proc Natl Acad Sci USA 102:5084–5089
Sambrook J, Russel DW (2001) Molecular cloning. In: A laboratory manual. 3rd edn. Cold Spring Harbor Laboratory Press, New York
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Úbeda-Manzanaro M, Merlo MA, Palazón JL, Cross I, Sarasquete C, Rebordinos L (2010) Chromosomal mapping of the major and minor ribosomal genes, (GATA)n and U2 snRNA gene by double-colour FISH in species of the Batrachoididae family. Genetica 138:787–794
Vierna J, Jensen KT, Martínez-Lage A, González-Tizón AM (2011) The linked units of 5S rDNA and U1 snDNA of razor shells (Mollusca: Bivalvia: Pharidae). Heredity 107:127–142
Vierna J, Wehner S, Zu Siederdissen CH, Martínez-Lage A, Marz M (2013) Systematic analysis and evolution of 5S ribosomal DNA in metazoans. Heredity 111:410–421
Vizoso M, Vierna J, González-Tizón AM, Martínez-Lage A (2011) The 5S rDNA gene family in Mollusks: characterization of transcriptional regulatory regions, prediction of secondary structures, and long-term evolution, with special attention to Mytilidae mussels. J Heredity 102:433–447
Wasko PA, Martins C, Wright JM, Galetti PM Jr (2001) Molecular organization of 5S rDNA in fishes of the genus Brycon. Genome 44:893–902
Acknowledgments
The authors are grateful to the “Parque Estadual Edmundo Navarro de Andrade” administration for sample collecting authorization and to the two anonymous reviewers for valuable suggestions. This study was partially supported by the Fundação de Amparo a Pesquisa do Estado de São Paulo-FAPESP (process numbers 2011/19481-3 and 2014/11763-8) and Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior-CAPES. DB and OMPG acknowledge the scholarships obtained from FAPESP (process numbers 2011/18028-3 and 2014/02038-8, respectively). DAM was supported by Consejo Nacional de Investigaciones Científicas y Técnicas-CONICET from Argentina. DCCM was the recipient of a research productivity fellowship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq (process number 304758/2014-0).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was founded by Fundação de Amparo a Pesquisa do Estado de São Paulo-FAPESP (process numbers 2011/19481-3 and 2014/11763-8) and Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior-CAPES.
Conflict of interest
Danilo Bueno declares that he has no conflict of interest. Octavio Manuel Palacios-Gimenez declares that he has no conflict of interest. Dardo Andrea Martí declares that he has no conflict of interest. Tatiane Casagrande Mariguela declares that she has no conflict of interest. Diogo Cavalcanti Cabral-de-Mello declares that he has no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Communicated by S. Hohmann.
D. Bueno and O. M. Palacios-Gimenez equally contributed.
Rights and permissions
About this article
Cite this article
Bueno, D., Palacios-Gimenez, O.M., Martí, D.A. et al. The 5S rDNA in two Abracris grasshoppers (Ommatolampidinae: Acrididae): molecular and chromosomal organization. Mol Genet Genomics 291, 1607–1613 (2016). https://doi.org/10.1007/s00438-016-1204-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-016-1204-1