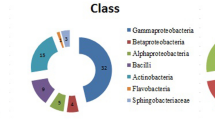
Abstract
Bacillus altitudinis is a widely distributed soil bacterium that has various functional activities, including remediation of contaminated soil, degradation of herbicides, and enhancement of plant growth. B. altitudinis GQYP101 was isolated from the rhizosphere soil of Lycium barbarum L. and demonstrated potential as a plant growth-promoting bacterium. In this work, strain GQYP101 could solubilize phosphorus, and increased the stem diameter, maximum leaf area, and fresh weight of corn in a pot experiment. Nitrogen and phosphorus contents of corn seedlings (aerial part) increased by 100% and 47.9%, respectively, after application of strain GQYP101. Concurrently, nitrogen and phosphorus contents of corn root also increased, by 55.40% and 20.3%, respectively. Furthermore, rhizosphere soil nutrients were altered and the content of available phosphorus increased by 73.2% after application of strain GQYP101. The mechanism by which strain GQYP101 improved plant growth was further investigated by whole genome sequence analysis. Strain GQYP101 comprises a circular chromosome and a linear plasmid. Some key genes of strain GQYP101 were identified that were related to phosphate solubilization, alkaline phosphatase, chemotaxis, and motility. The findings of this study may provide a theoretical basis for strain GQYP101 to enhance crop yield as microbial fertilizer.




Similar content being viewed by others
Data Availability
All data of this study are available within the manuscript and supplementary materials.
References
Backer R, Rokem JS, Ilangumaran G, Lamont J, Praslickova D, Ricci E, Subramanian S, Smith DL (2018) Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front Plant Sci 9:1473. https://doi.org/10.3389/fpls.2018.01473
You M, Fang S, MacDonald J, Xu J, Yuan Z-C (2020) Isolation and characterization of Burkholderia cenocepacia CR318, a phosphate solubilizing bacterium promoting corn growth. Microbiol Res 233:126395. https://doi.org/10.1016/j.micres.2019.126395
Yue Z, Shen Y, Chen Y, Liang A, Chu C, Chen C, Sun Z (2019) Microbiological insights into the stress-alleviating property of an endophytic Bacillus altitudinis WR10 in wheat under low-phosphorus and high-salinity stresses. Microorganisms 7:508. https://doi.org/10.3390/microorganisms7110508
Narayanasamy S, Thangappan S, Uthandi S (2020) Plant growth-promoting Bacillus sp cahoots moisture stress alleviation in rice genotypes by triggering antioxidant defense system. Microbiol Res 239:126518. https://doi.org/10.1016/j.micres.2020.126518
Pereira S, Abreu D, Moreira H, Vega A, Castro P (2020) Plant growth-promoting rhizobacteria (PGPR) improve the growth and nutrient use efficiency in maize (Zea mays L) under water deficit conditions. Heliyon 6:e05106. https://doi.org/10.1016/j.heliyon.2020.e05106
Zhang Y, Chen C-X, Feng H-P, Wang X-J, Roessner U, Walker R, Cheng Z-Y, An Y-Q, Du B, Bai J-G (2020) Transcriptome profiling combined with activities of antioxidant and soil enzymes reveals an ability of Pseudomonas sp CFA to mitigate p-hydroxybenzoic and ferulic acid stresses in cucumber. Front Microbiol 11:2706. https://doi.org/10.3389/fmicb.2020.522986
Pranaw K, Pidlisnyuk V, Trögl J, Malinská H (2020) Bioprospecting of a novel plant growth-promoting bacterium Bacillus altitudinis KP-14 for enhancing Miscanthus × giganteus growth in metals contaminated soil. Biology 9:305. https://doi.org/10.3390/biology9090305
Wang C, Zhao D, Qi G, Mao Z, Hu X, Du B, Liu K, Ding Y (2020) Effects of Bacillus velezensis FKM10 for promoting the growth of Malus hupehensis Rehd and inhibiting Fusarium verticillioides. Front Microbiol 10:2889. https://doi.org/10.3389/fmicb.2019.02889
Bhattacharyya PN, Jha DK (2012) Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol 28:1327–1350. https://doi.org/10.1007/s11274-011-0979-9
Shivaji S, Chaturvedi P, Suresh K, Reddy G, Dutt C, Wainwright M, Narlikar JV, Bhargava P (2006) Bacillus aerius sp. nov., Bacillus aerophilus sp. nov., Bacillus stratosphericus sp. nov. and Bacillus altitudinis sp. nov., isolated from cryogenic tubes used for collecting air samples from high altitudes. Int J Syst Evol Microbiol 56:1465–1473. https://doi.org/10.1099/ijs.0.64029-0
Kaur R, Goyal D (2020) Biodegradation of butachlor by Bacillus altitudinis and identification of metabolites. Current Microbiol 77:2602–2612. https://doi.org/10.1007/s00284-020-02031-1
Yue Z, Chen Y, Chen C, Ma K, Tian E, Wang Y, Liu H, Sun Z (2021) Endophytic Bacillus altitudinis WR10 alleviates Cu toxicity in wheat by augmenting reactive oxygen species scavenging and phenylpropanoid biosynthesis. J Hazard Mater 405:124272. https://doi.org/10.1016/j.jhazmat.2020.124272
Goswami M, Deka S (2019) Biosurfactant production by a rhizosphere bacteria Bacillus altitudinis MS16 and its promising emulsification and antifungal activity. Colloids Surf B Biointerfaces 178:285–296. https://doi.org/10.1016/j.colsurfb.2019.03.003
Othman R, Panhwar QA (2015) Phosphate-solubilizing bacteria improves nutrient uptake in aerobic rice. In: Khan MS, Zaidi A, Musarrat J (eds) Phosphate solubilizing microorganisms. Springer, Cham, pp 207–224
Pande A, Pandey P, Mehra S, Singh M, Kaushik S (2017) Phenotypic and genotypic characterization of phosphate solubilizing bacteria and their efficiency on the growth of maize. J Genet Eng Biotechnol 15:379–391. https://doi.org/10.1016/j.jgeb.2017.06.005
Xue H-P, Zhang D-F, Xu L, Wang X-N, Zhang A-H, Huang J-K, Liu C (2021) Actirhodobacter atriluteus gen. nov., sp. Nov., isolated from the surface water of the Yellow Sea. Antonie van Leeuwenhoek 114:1–10. https://doi.org/10.1007/s10482-021-01576-w
Nautiyal CS (1999) An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett 170:265–270. https://doi.org/10.1111/j.1574-6968.1999.tb13383.x
Bowman R (1988) A rapid method to determine total phosphorus in soils. Soil Sci Soc Am J 52:1301–1304. https://doi.org/10.2136/sssaj1988.03615995005200050016x
Yu C, Hu X, Deng W, Li Y, Xiong C, Ye C, Han G, Li X (2015) Changes in soil microbial community structure and functional diversity in the rhizosphere surrounding mulberry subjected to long-term fertilization. Appl Soil Ecol 86:30–40. https://doi.org/10.1016/j.apsoil.2014.09.013
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Ma J, Wang C, Wang H, Liu K, Zhang T, Yao L, Zhao Z, Du B, Ding Y (2018) Analysis of the complete genome sequence of Bacillus atrophaeus GQJK17 reveals its biocontrol characteristics as a plant growth-promoting rhizobacterium. Biomed Res Int. https://doi.org/10.1155/2018/9473542
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. https://doi.org/10.1093/nar/25.5.955
Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A (2013) Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 41:D226–D232. https://doi.org/10.1093/nar/gks1005
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. https://doi.org/10.1093/nar/29.12.2607
Krogh A, Brown M, Mian IS, Sjölander K, Haussler D (1994) Hidden Markov models in computational biology: applications to protein modeling. J Mol Biol 235:1501–1531. https://doi.org/10.1006/jmbi.1994.1104
Petersen TN, Brunak S, Von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat methods 8:785–786. https://doi.org/10.1038/nmeth.1701
Chen Y, Yu P, Luo J, Jiang Y (2003) Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm Genome 14:859–865. https://doi.org/10.1007/s00335-003-2296-6
Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, Von Mering C, Bork P (2017) Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34:2115–2122. https://doi.org/10.1093/molbev/msx148
Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang H-Y, Dosztányi Z, El-Gebali S, Fraser M (2017) InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res 45:D190–D199. https://doi.org/10.1093/nar/gkw1107
Lam MQ, Chen SJ, Goh KM, Abd Manan F, Yahya A, Shamsir MS, Chong CS (2020) Genome sequence of an uncharted halophilic bacterium Robertkochia marina with deciphering its phosphate-solubilizing ability. Braz J Microbiol. https://doi.org/10.1007/s42770-020-00401-2
Arcondéguy T, Jack R, Merrick M (2001) PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol Mol Biol Rev 65:80–105. https://doi.org/10.1128/MMBR.65.1.80-105.2001
Nannan C, Vu HQ, Gillis A, Caulier S, Mahillon J (2020) Bacilysin within the Bacillus subtilis group: gene prevalence versus antagonistic activity against gram-negative foodborne pathogens. J Biotechnol 327:28–35. https://doi.org/10.1016/j.jbiotec.2020.12.017
Schalk IJ, Rigouin C, Godet J (2020) An overview of siderophore biosynthesis among fluorescent Pseudomonads and new insights into their complex cellular organization. Environ Microbiol 22:1447–1466. https://doi.org/10.1111/1462-2920.14937
Ryu CM, Farag MA, Hu CH, Reddy MS, Kloepper JW (2003) Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci USA 100:4927–4932. https://doi.org/10.1073/pnas.0730845100
Cesari A, Paulucci N, López-Gómez M, Hidalgo-Castellanos J, Plá CL, Dardanelli MS (2019) Restrictive water condition modifies the root exudates composition during peanut-PGPR interaction and conditions early events, reversing the negative effects on plant growth. Plant Physiol Biochem 142:519–527. https://doi.org/10.1016/j.plaphy.2019.08.015
Potshangbam M, Sahoo D, Verma P, Verma S, Kalita MC, Devi SI (2018) Draft genome sequence of Bacillus altitudinis Lc5, a biocontrol and plant growth-promoting endophyte strain isolated from indigenous black rice of Manipur. Genome Announc. https://doi.org/10.1128/genomeA.00601-18
Ahemad M (2015) Phosphate-solubilizing bacteria-assisted phytoremediation of metalliferous soils: a review. 3 Biotech 5:111–121. https://doi.org/10.1007/s13205-014-0206-0
Billah M, Khan M, Bano A, Hassan TU, Munir A, Gurmani AR (2019) Phosphorus and phosphate solubilizing bacteria: keys for sustainable agriculture. Geomicrobiol J 36:904–916. https://doi.org/10.1080/01490451.2019.1654043
Rodríguez H, Fraga R, Gonzalez T, Bashan Y (2006) Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. Plant Soil 287:15–21. https://doi.org/10.1007/s11104-006-9056-9
Suleman M, Yasmin S, Rasul M, Yahya M, Atta BM, Mirza MS (2018) Phosphate solubilizing bacteria with glucose dehydrogenase gene for phosphorus uptake and beneficial effects on wheat. PLoS ONE 13:e0204408. https://doi.org/10.1371/journal.pone.0204408
Tang Z, Zhang L, He N, Gong D, Gao H, Ma Z, Fu L, Zhao M, Wang H, Wang C (2021) Soil bacterial community as impacted by addition of rice straw and biochar. Sci Rep-UK 11(1):1–9. https://doi.org/10.1038/s41598-021-99001-9
Zhang W, Chen L, Zhang R, Lin K (2016) High throughput sequencing analysis of the joint effects of BDE209-Pb on soil bacterial community structure. J Hazard Mater 301:1–7. https://doi.org/10.1016/j.jhazmat.2015.08.037
Felici C, Vettori L, Giraldi E, Forino LMC, Toffanin A, Tagliasacchi AM, Nuti M (2008) Single and co-inoculation of Bacillus subtilis and Azospirillum brasilense on Lycopersicon esculentum: effects on plant growth and rhizosphere microbial community. Appl Soil Ecol 40:260–270. https://doi.org/10.1016/j.apsoil.2008.05.002
Chen T, Hu R, Zheng Z, Yang J, Fan H, Deng X, Yao W, Wang Q, Peng S, Li J (2021) Soil bacterial community in the multiple cropping system increased grain yield within 40 cultivation years. Front Plant Sci. https://doi.org/10.3389/fpls.2021.804527
Liu C, Dong Y, Hou L, Deng N, Jiao R (2017) Acidobacteria community responses to nitrogen dose and form in chinese fir plantations in Southern China. Curr Microbiol 74(3):396–403. https://doi.org/10.1007/s00284-016-1192-8
Zhelezova A, Chernov T, Tkhakakhova A, Xenofontova N, Semenov M, Kutovaya O (2019) Prokaryotic community shifts during soil formation on sands in the tundra zone. PLoS ONE 14(4):e0206777. https://doi.org/10.1371/journal.pone.0206777
Zhang L, Xu M, Liu Y, Zhang F, Hodge A, Feng G (2016) Carbon and phosphorus exchange may enable cooperation between an arbuscular mycorrhizal fungus and a phosphate-solubilizing bacterium. New Phytol 210:1022–1032. https://doi.org/10.1111/nph.13838
Zhang L, Feng G, Declerck S (2018) Signal beyond nutrient, fructose, exuded by an arbuscular mycorrhizal fungus triggers phytate mineralization by a phosphate solubilizing bacterium. ISME J 12:2339–2351. https://doi.org/10.1038/s41396-018-0171-4
Funding
The National Natural Science Foundation of China (31700094) supported Dr. Chengqiang Wang. The National Natural Science Foundation of China (31770115) supported Dr. Yanqin Ding. The National Key Research and Development Program of China (No. 2017YFD0200804), the Science and Technology Project of Guizhou (ZYJ2017-8), and the Key Field Research and Development Program of Guangdong Province (2019B020218009) supported Prof. Binghai Du.
Author information
Authors and Affiliations
Contributions
DZ and YC performed the work and analyzed the data. DZ and YD wrote the original draft. CW revised the manuscript. YZ, KL, LY, XH, YP, and JG advised the manuscript. CW and BD supported the study.
Corresponding author
Ethics declarations
Conflict of Interest
All the authors declare no competing interests.
Ethical Approval
The research does not involve human and animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, D., Ding, Y., Cui, Y. et al. Isolation and Genome Sequence of a Novel Phosphate-Solubilizing Rhizobacterium Bacillus altitudinis GQYP101 and Its Effects on Rhizosphere Microbial Community Structure and Functional Traits of Corn Seedling. Curr Microbiol 79, 249 (2022). https://doi.org/10.1007/s00284-022-02944-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-02944-z