Abstract
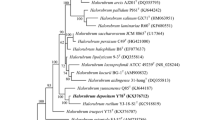
Brevibacterium halotolerans is currently classified as a member of the Brevibacterium genus, a genus that groups together many bacterial species of similar morphology but diverse biochemical and physiological features. Here we suggest, based on multiple gene sequencing and microbial and biochemical characterization of two environmental isolates and one type strain (DSM8802), that the B. halotolerans DSM8802 (and probably the other deposited under this species name) should be re-classified into the Bacillus genus, and offered the name B. halotolerans comb. nov.



Similar content being viewed by others
References
Abou-Shanab RK, Ghanem N, Ghanem N, Al-Kolaibe A (2008) The role of bacteria on heavy-metal extraction and uptake by plants growing on multi-metal-contaminated soils. World J Microbiol Biotechnol 24:253–262
Abusham RA, Rahman RN, Salleh AB, Basri M (2009) Optimization of physical factors affecting the production of thermo-stable organic solvent-tolerant protease from a newly isolated halo tolerant Bacillus subtilis strain Rand. Microb Cell Fact 9:8–20
Acharya KP, Shilpkar M, Shah MC, Chellapandi P (2015) Biodegradation of insecticide monocrotophos by Bacillus subtilis KPA-1, isolated from agriculture soils. Appl Biochem Biotechnol 175:1789–1804
Bacon CW, Hinton DM (2002) Endophytic and biological control potential of Bacillus mojavensis and related species. Biol Control 23:274–284
Breed RS (1953) The Brevibacteriaceae fam. nov. of order Eubacteriales. Rias Commun VI Congr Int Microbiol Roma 1:10–15
Collins MD (2006) The genus Brevibacterium. The Prokaryotes. Springer, New York, pp 1013–1019
Delaporte B, Sasson A (1967) [Study of bacteria from arid soils of Morocco: Bacillus maroccanus n. sp]. Comptes Rendus Hebdomadaires des Seances de L’Academie des Sciences. Serie D 264(18):2257–2260
DNA Baser sequence assembler v4 (2013) Heracle BioSoft, www.DnaBaser.com
DSM webpage—Comment on DSM 8802 Comment: not a Brevibacterium; affiliated to the Bacillus mojavensis cluster. Retrieved from https://www.dsmzde/catalogues/details/culture/dsm-8802.html on April 18, 2016; See also https://www.dsmz.de/microorganisms/wink_pdf/DSM8802.pdf
Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans AD (1997) Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology 143(9):2983–2989
Gelsomino R, Vancanneyt M, Vandekerckhove TM, Swings J (2004) Development of a 16S rRNA primer for the detection of Brevibacterium spp. Lett Appl Microbiol 38:532–535
Gerhardt P (1994) Methods for general and molecular bacteriology. American Society for Microbiology Press, Washington, DC
Hayyat R, Khalidi R, Ehsani M, Ahmed I, Yokota A, Ali S (2013) Molecular characterization of soil bacteria for improving crop yield in Pakistan. Pak J Bot 45(3):1045–1055
Jones D, Keddie RM (1985) Genus Brevibacterium. In: Sheath PHA, Mair NA, Shape ME, Glott JG (eds) Bergey’s manual of systematic bacteriology, vol vol 2. Springer, New York, pp 1301–1313
Jurado M, López MJ, Suárez-Estrella F, Vargas-García MC, López-González JA, Moreno J (2014) Exploiting composting biodiversity: study of the persistent and biotechnologically relevant microorganisms from lignocellulose-based composting. Bioresour Technol 162:283–293
Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A (2008) A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr Microbiol 57:503–507
Khan MN, Lin H, Wang J, Li M, Mirani ZA (2014) recA- based identification and antagonistic potential against fish pathogens by marine isolate DK1-SA11. Indian J Geo Marine Sci 43:731–736
Kim O, Cho Y, Lee K, Yoon S, Kim M, Na H, Park S, Jeon Y, Lee J, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62(3):716–721
Roberts MS, Nakamura LK, Cohan FM (1994) Bacillus mojavensis sp. nov., distinguishable from Bacillus subtilis by sexual isolation, divergence in DNA sequence, and differences in fatty acid composition. Int J Syst Bacteriol 44:256–264
Schaeffer AB, Fulton M (1933) A simplified method of staining endospores. Science 77:194
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular Evolutionary Genetics Analysis version 60. Mol Biol Evol 30:2725–2729
Acknowledgments
The authors want to acknowledge Aharon Oren for his useful discussion and for suggesting the new name. This study was funded by the Israeli Ministry of Environmental Protection Grant Number 122-3-2. We also wish to acknowledge the reviewers for helpful comments.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ben-Gad, D., Gerchman, Y. Reclassification of Brevibacterium halotolerans DSM8802 as Bacillus halotolerans comb. nov. Based on Microbial and Biochemical Characterization and Multiple Gene Sequence. Curr Microbiol 74, 1–5 (2017). https://doi.org/10.1007/s00284-016-1143-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-016-1143-4