Abstract
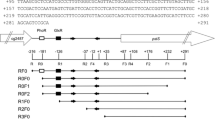
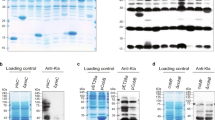
pEMBTL-SY1, which can over produce the ArgR protein in Corynebacterium glutamicum, was constructed. The DNA-binding affinity of ArgR was analyzed using a Chromatin Immunoprecipitation (ChIP) assay. The level of ArgR protein expression in the plasmid-carrying C. glutamicum (pEMBTL-SY1) was higher than that in the wild-type strain. On the other hand, there was no increase in the DNA-binding affinity of ArgR on the upstream of argB and the level of ornithine production. The DNA-binding affinity of ArgR on the arg operon and the level of ornithine production in the presence of three metabolites, ornithine, arginine, and proline, were examined as feedback controlling effectors in the arginine biosynthesis pathway in C. glutamicum. The ChIP assay showed that the supplemented metabolites altered the ArgR-binding affinity on the upstream of argB, which is consistent with the change in ornithine production. This suggests that the regulation of ornithine biosynthesis by the transcriptional regulator, ArgR, depends on the DNA-binding affinity of the arg operon, which is regulated by the feedback controlling effectors, rather than on the level of ArgR protein expression.




Similar content being viewed by others
References
Caldara M, Minh PNL, Bostoen S et al (2007) ArgR-dependent repression of arginine and histidine transport genes in Escherichia coli K-12. J Mol Biol 373:251–267
Caldra M, Charlier D, Cunin R (2006) The arginine regulon of Escherichia coli: whole-system transcriptome analysis discover new genes and provides an integrated view of arginine regulon. Microbiology 152:3343–3354
Chinard FP (1952) Photometric estimation of proline and ornithine. J Biol Chem 199:91–95
Cunin P, Glansdorff N, Piérard A et al (1986) Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev 50:314–352
Dubois E, Messenguy F (1991) In vitro studies of the binding of the ARGR protein to the ARG5, 6 promoter. Mol Cell Biol 11:2162–2168
Eikmanns BJ, Thus-Schmitz N, Eggeling L et al (1994) Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene coding citrate synthase. Microbiology 140:1817–1828
Fernández-Murga ML, Gil-Ortiz F, Llácer JL et al (2004) Arginine biosynthesis in Thermotoga maritima: characterization of the arginine-sensitive N-acetyl-l-glutamate kinase. J Bacteriol 186:6142–6149
Gallegos M-T, Schleif R, Bairoch A et al (1997) AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev 61:393–410
Grandori R, Lavoie TA, Pflumm M et al (1995) The DNA-binding domain of the hexameric arginine repressor. J Mol Biol 254:150–162
Graupner M, White RH (2001) Methanococcus jannaschii generates L-proline by cyclization of L-ornithine. J Bacteriol 183:5203–5205
Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Bacteriol 104:155–172
Hwang J-H, Hwang G-H, Cho J-Y (2008) Effect of increased glutamate availability on l-ornithine production in Corynebacterium glutamicum. J Microbiol Biotechnol 18:704–710
Kiupakis AK, Reitzer L (2002) ArgR-independent induction and ArgR-dependent superinduction of the astCADBE operon in Escherichia coli. J Bacteriol 184:2940–2950
Larsen R, Kok J, Kuipers OP (2005) Interaction between ArgR and AhrC controls regulation of arginine metabolism in Lactococcus lactis. J Biol Chem 280:19319–19330
Lee H-W, Yoon S-J, Jang H-W et al (2000) Effect of mixing on fed-batch fermentation of l-ornithine. J Biosci Bioeng 89:539–544
Lee Y-J, Cho J-Y (2006) Genetic manipulation of a primary metabolic pathway for l-ornithine production in Escherichia coli. Biotechnol Lett 28:1849–1856
Liebl W, Bayerl A, Schein B et al (1989) High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol Lett 65:299–304
Lu C-D (2006) Pathways and regulation of bacterial arginine metabolism and perspectives for obtaining arginine overproducing strains. Appl Microbiol Biotechnol 70:261–272
Lu C-D, Abdelal AT (1999) Role of ArgR in activation of the ast operon, encoding enzymes of the arginine succinyltransferase pathway in Salmonella typhimurium. J Bacteriol 181:1934–1938
Maas WK (1994) The arginine repressor of Escherichia coli. Microbiol Rev 58:631–640
Nishijyo T, Park S-M, Lu C-D et al (1998) Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J Bacteriol 180:5559–5566
Sakanyan V, Petrosyan P, Lecocq M et al (1996) Genes and enzymes of the acetyl cycle of arginine biosynthesis in Corynebacterium glutamicum: enzyme evolution in the early steps of the arginine pathway. Microbiology 142:99–108
Salvatore F, Cimino F, Maria C et al (1964) Mechanism of the protection by l-ornithine-l-aspartate mixture and by l-arginine in ammonia intoxication. Arch Biochem Biophys 107:499–503
Sambrook J, Fritsch EF, Maniatis T (2001) Molecular cloning: a laboratory manual, vol 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Udaka S (1960) Screening method for microorganisms accumulating metabolites and its use in the isolation of Micrococcus glutamicus. J Bacteriol 79:754–755
Xu Y, Sun Y, Huysveld N et al (2003) Regulation of arginine biosynthesis in the psychropiezophilic bacterium Moritella profunda: in vivo repressibility and in vitro repressor-operator contact probing. J Mol Biol 326:353–369
Xu Y, Labedan B, Glansdorff N (2007) Surprising arginine biosynthesis: a reappraisal of the enzymology and evolution of the pathway in microorganisms. Microbiol Mol Biol Rev 71:36–47
Acknowledgments
This study was supported by the 21C Frontier Microbial Genomics and Applications Center Program, Grant No. 11-2008-10-002-00, Ministry of Education, Science & Technology, Republic of Korea. Further support from a 2008 research grant from Chungbuk National University is appreciated. The authors are grateful for their supports.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Lee, S.Y., Kim, YH. & Min, J. The Effect of ArgR–DNA Binding Affinity on Ornithine Production in Corynebacterium glutamicum . Curr Microbiol 59, 483–488 (2009). https://doi.org/10.1007/s00284-009-9467-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-009-9467-y