Abstract
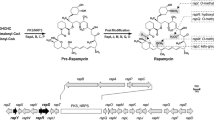
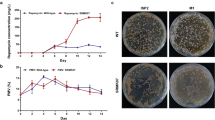
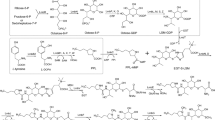
Rapamycin is an important macrocyclic antibiotic produced by Streptomyces rapamycinicus. In the rapamycin biosynthetic gene cluster (BGC), there are up to five regulatory genes, which have been shown to play important roles in the regulation of rapamycin biosynthesis. Here, we demonstrated that the rapamycin BGC-situated LAL family regulator RapH co-ordinately regulated the biosynthesis of both rapamycin and elaiophylin. We showed that rapH overexpression not only resulted in enhanced rapamycin production but also led to increased synthesis of another type I polyketide antibiotic, elaiophylin. Consistent with this, rapH deletion resulted in decreased production of both antibiotics. Through real-time RT-PCR combined with β-glucuronidase reporter assays, four target genes controlled by RapH, including rapL (encoding a lysine cyclodeaminase)/rapH in the rapamycin BGC and ela3 (encoding a LuxR family regulator)/ela9 (encoding a hypothetical protein) in the elaiophylin BGC, were identified. A relatively conserved signature sequence recognized by RapH, which comprises two 4-nt inverted repeats separated by 8-nt, 5′-GTT/AC-N8-GTAC-3′, was defined. Taken together, our findings demonstrated that RapH was involved in co-ordinated regulation of two disparate BGCs specifying two unrelated antibiotics, rapamycin and elaiophylin. These results further expand our knowledge of the regulation of antibiotic biosynthesis in S. rapamycinicus.
Key points
• The cluster-situated regulator RapH controlled the synthesis of two antibiotics.
• Four promoter regions recognized by RapH were identified.
• A 16-nt signature DNA sequence essential for RapH regulation was defined.






Similar content being viewed by others
References
Andexer JN, Kendrew SG, Nur-e-Alam M, Lazos O, Foster TA, Zimmermann AS, Warneck TD, Suthar D, Coates NJ, Koehn FE, Skotnicki JS, Carter GT, Gregory MA, Martin CJ, Moss SJ, Leadlay PF, Wilkinson B (2011) Biosynthesis of the immunosuppressants FK506, FK520, and rapamycin involves a previously undescribed family of enzymes acting on chorismate. Proc Natl Acad Sci U S A 108:4776–4781
Aparicio JF, Molnar I, Schwecke T, Konig A, Haydock SF, Khaw LE, Staunton J, Leadlay PF (1996) Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene 169:9–16
Argyriou P, Economopoulou P, Papageorgiou S (2012) The Role of mTOR inhibitors for the treatment of B-Cell lymphomas. Adv Hematol 2012:435342
Barreales EG, Vicente CM, de Pedro A, Santos-Aberturas J, Aparicio JF (2018) Promoter engineering reveals the importance of heptameric direct repeats for DNA binding by streptomyces antibiotic regulatory protein-large ATP-binding regulator of the LuxR family (SARP-LAL) regulators in Streptomyces natalensis. Appl Environ Microbiol 84:e00246
Chung L, Liu L, Patel S, Carney JR, Reeves CD (2001) Deletion of rapQONML from the rapamycin gene cluster of Streptomyces hygroscopicus gives production of the 16-O-desmethyl-27-desmethoxy analog. J Antibiot (tokyo) 54:250–256
Fang A, Wong GK, Demain AL (2000) Enhancement of the antifungal activity of rapamycin by the coproduced elaiophylin and nigericin. J Antibiot (tokyo) 53:158–162
Gomez-Escribano JP, Bibb MJ (2011) Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol 4:207–215
Gregory MA, Gaisser S, Lill RE, Hong H, Sheridan RM, Wilkinson B, Petkovic H, Weston AJ, Carletti I, Lee HL, Staunton J, Leadlay PF (2004) Isolation and characterization of pre-rapamycin, the first macrocyclic intermediate in the biosynthesis of the immunosuppressant rapamycin by S. hygroscopicus. Angew Chem Int Ed Engl 43:2551–2553
Guan H, Li Y, Zheng J, Liu N, Zhang J, Tan H (2019) Important role of a LAL regulator StaR in the staurosporine biosynthesis and high-production of Streptomyces fradiae CGMCC 4.576. Sci China Life Sci 62:1638–1654
Gui M, Zhang MX, Wu WH, Sun P (2019) Natural occurrence, bioactivity and biosynthesis of elaiophylin analogues. Molecules 24:3840
Guo J, Zhao JL, Li LL, Chen Z, Wen Y, Li JL (2010) The pathway-specific regulator AveR from Streptomyces avermitilis positively regulates avermectin production while it negatively affects oligomycin biosynthesis. Mol Genet Genomics 283:123–133
Hamedi J, Mohammadipanah F, Klenk HP, Potter G, Schumann P, Sproer C, von Jan M, Kroppenstedt RM (2010) Streptomyces iranensis sp. nov., isolated from soil. Int J Syst Evol Microbiol 60:1504–1509
Hammann P, Kretzschmar G, Seibert G (1990) Secondary metabolites by chemical screening. 7. I. Elaiophylin derivatives and their biological activities. J Antibiot (tokyo) 43:1431–1440
Haydock SF, Mironenko T, Ghoorahoo HI, Leadlay PF (2004) The putative elaiophylin biosynthetic gene cluster in Streptomyces sp. DSM4137 is adjacent to genes encoding adenosylcobalamin-dependent methylmalonyl CoA mutase and to genes for synthesis of cobalamin. J Biotechnol 113:55–68
Horn F, Schroeckh V, Netzker T, Guthke R, Brakhage AA, Linde J (2014) Draft genome sequence of Streptomyces iranensis. Genome Announc 2:e00616
Houchens DP, Ovejera AA, Riblet SM, Slagel DE (1983) Human brain tumor xenografts in nude mice as a chemotherapy model. Eur J Cancer Clin Oncol 19:799–805
Huang H, Ren SX, Yang S, Hu HF (2015a) Comparative analysis of rapamycin biosynthesis clusters between Actinoplanes sp. N902–109 and Streptomyces hygroscopicus ATCC29253. Chin J Nat Med 13:90–98
Huang H, Zheng GS, Jiang WH, Hu HF, Lu YH (2015b) One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Bioch Bioph Sin 47:231–243
Jiang M, Yin M, Wu S, Han X, Ji K, Wen M, Lu T (2017) GdmRIII, a TetR Family Transcriptional Regulator, Controls Geldanamycin and Elaiophylin Biosynthesis in Streptomyces autolyticus CGMCC0516. Sci Rep 7:4803
Kieser T, Bibb MJ, Buttner MJ, Chater KF (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich, England
Klassen JL, Lee SR, Poulsen M, Beemelmanns C, Kim KH (2019) Efomycins K and L from a termite-associated Streptomyces sp. M56 and their putative biosynthetic origin. Front Microbiol 10:1739
Konig A, Schwecke T, Molnar I, Bohm GA, Lowden PA, Staunton J, Leadlay PF (1997) The pipecolate-incorporating enzyme for the biosynthesis of the immunosuppressant rapamycin-nucleotide sequence analysis, disruption and heterologous expression of rapP from Streptomyces hygroscopicus. Eur J Biochem 247:526–534
Kuscer E, Coates N, Challis I, Gregory M, Wilkinson B, Sheridan R, Petkovic H (2007) Roles of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J Bacteriol 189:4756–4763
Li C, He H, Wang J, Liu H, Wang H, Zhu Y, Wang X, Zhang Y, Xiang W (2019) Characterization of a LAL-type regulator NemR in nemadectin biosynthesis and its application for increasing nemadectin production in Streptomyces cyaneogriseus. Sci China Life Sci 62:394–405
Liu H, Naismith JH (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 8:91
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408
Malagelada C, Jin ZH, Jackson-Lewis V, Przedborski S, Greene LA (2010) Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson’s disease. J Neurosci 30:1166–1175
Martel RR, Klicius J, Galet S (1977) Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Can J Physiol Pharmacol 55:48–51
McLean TC, Hoskisson PA, Seipke RF (2016) Coordinate regulation of antimycin and candicidin biosynthesis. mSphere 1:e00305
McLean TC, Wilkinson B, Hutchings MI, Devine R (2019) Dissolution of the disparate: co-ordinate regulation in antibiotic biosynthesis. Antibiotics (basel) 8:83
Myronovskyi M, Welle E, Fedorenko V, Luzhetskyy A (2011) Beta-glucuronidase as a sensitive and versatile reporter in Actinomycetes. Appl Environ Microbiol 77:5370–5383
Paiva NL, Demain AL, Roberts MF (1991) Incorporation of acetate, propionate, and methionine into rapamycin by Streptomyces hygroscopicus. J Nat Prod 54:167–177
Pan T, Kondo S, Zhu W, Xie W, Jankovic J, Le W (2008) Neuroprotection of rapamycin in lactacystin-induced neurodegeneration via autophagy enhancement. Neurobiol Dis 32:16–25
Park SR, Yoo YJ, Ban YH, Yoon YJ (2010) Biosynthesis of rapamycin and its regulation: past achievements and recent progress. J Antibiot (tokyo) 63:434–441
Schrijver A, Mot R (1999) A subfamily of MalT-related ATP-dependent regulators in the LuxR family. Microbiology (reading) 145(Pt 6):1287–1288
Schwecke T, Aparicio JF, Molnar I, Konig A, Khaw LE, Haydock SF, Oliynyk M, Caffrey P, Cortes J, Lester JB (1995) The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci U S A 92:7839–7843
Strohl WR (1992) Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res 20:961–974
Tain LS, Mortiboys H, Tao RN, Ziviani E, Bandmann O, Whitworth AJ (2009) Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci 12:1129–1135
Vezina C, Kudelski A, Sehgal SN (1975) Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (tokyo) 28:721–726
Wilkinson CJ, Hughes-Thomas ZA, Martin CJ, Bohm I, Mironenko T, Deacon M, Wheatcroft M, Wirtz G, Staunton J, Leadlay PF (2002) Increasing the efficiency of heterologous promoters in actinomycetes. J Mol Microbiol 4:417–426
Wu C, Tan Y, Gan M, Wang Y, Guan Y, Hu X, Zhou H, Shang X, You X, Yang Z, Xiao C (2013) Identification of elaiophylin derivatives from the marine-derived actinomycete Streptomyces sp. 7–145 using PCR-based screening. J Nat Prod 76:2153–2157
Yoo YJ, Hwang JY, Shin H, Cui H, Lee J, Yoon YJ (2015) Characterization of negative regulatory genes for the biosynthesis of rapamycin in Streptomyces rapamycinicus and its application for improved production. J Ind Microbiol Biotechnol 42:125–135
Yoo YJ, Kim H, Park SR, Yoon YJ (2017) An overview of rapamycin: from discovery to future perspectives. J Ind Microbiol Biotechnol 44:537–553
Zhang YY, He HR, Liu H, Wang HY, Wang XJ, Xiang WS (2016) Characterization of a pathway-specific activator of milbemycin biosynthesis and improved milbemycin production by its overexpression in Streptomyces bingchenggensis. Microb Cell Fact 15:152
Zhao X, Fang Y, Yang Y, Qin Y, Wu P, Wang T, Lai H, Meng L, Wang D, Zheng Z, Lu X, Zhang H, Gao Q, Zhou J, Ma D (2015) Elaiophylin, a novel autophagy inhibitor, exerts antitumor activity as a single agent in ovarian cancer cells. Autophagy 11:1849–1863
Zhou HY, Huang SL (2012) Current development of the second generation of mTOR inhibitors as anticancer agents. Chin J Cancer 31:8–18
Zhu Z, Li H, Yu P, Guo Y, Luo S, Chen Z, Mao X, Guan W, Li Y (2017) SlnR is a positive pathway-specific regulator for salinomycin biosynthesis in Streptomyces albus. Appl Microbiol Biotechnol 101:1547–1557
Funding
This study was supported by the National Natural Science Foundation of China (31770088 and 31970083) and the National Key Research and Development Program (2019YFA0905400, 2021YFC2100600, and 2018YFA0903700).
Author information
Authors and Affiliations
Contributions
YHL and JZT conceived and designed the study. WYH, JXM, JZT, and GSZ performed the experiments. WWF, YHL, and WHJ supervised the experiments. AAZ performed the bioinformatics analysis. WYH and YHL wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare no competing interests..
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
He, W., Wang, W., Ma, J. et al. Crossregulation of rapamycin and elaiophylin biosynthesis by RapH in Streptomyces rapamycinicus. Appl Microbiol Biotechnol 106, 2147–2159 (2022). https://doi.org/10.1007/s00253-022-11847-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-11847-9