Abstract
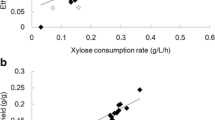
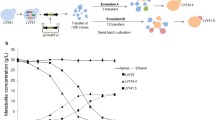
It is of utmost importance to construct industrial xylose-fermenting Saccharomyces cerevisiae strains for lignocellulosic bioethanol production. In this study, two xylose isomerase-based industrial S. cerevisiae strains, O7 and P5, were constructed by δ-integration of the xylose isomerase (XI) gene xylA from the fungus Orpinomyces sp. and from the bacterium Prevotella ruminicola, respectively. The xylose consumption of the strains O7 and P5 at 48-h fermentation was 17.71 and 26.10 g/L, respectively, in synthetic medium with xylose as the sole sugar source. Adaptive evolution further improved the xylose fermentation capacity of the two strains to 51.0 and 28.9% in average, respectively. The transcriptomes of these two strains before and after evolution were analyzed using RNA-Seq. The expression levels of the genes involved in cell integrity, non-optimal sugar utilization, and stress response to environment were significantly up-regulated after evolution and did not depend on the origin of xylA; the expression levels of the genes involved in transmembrane transport, rRNA processing, cytoplasmic translation, and other processes were down-regulated. The expression of genes involved in central carbon metabolism was fine-tuned after the evolution. The analysis of transcription factors (TFs) indicated that most of the genes with significant differential expression were regulated by the TFs related to cell division, DNA damage response, or non-optimal carbon source utilization. The results of this study could provide valuable references for the construction of efficient xylose-fermenting XI strains.





Similar content being viewed by others
References
Agarwala SD, Blitzblau HG, Hochwagen A, Fink GR (2012) RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet 8(6):e1002732
Brink DP, Borgström C, Tueros FG, Gorwa-Grauslund MF (2016) Real-time monitoring of the sugar sensing in Saccharomyces cerevisiae indicates endogenous mechanisms for xylose signaling. Microb Cell Factories 15:183
Buijs NA, Siewers V, Nielsen J (2013) Advanced biofuel production by the yeast Saccharomyces cerevisiae. Curr Opin Chem Biol 17(3):480–488
Demeke MM, Dietz H, Li Y, Foulquié-Moreno MR, Mutturi S, Deprez S, Abt TD, Bonini BM, Lidén G, Dumortier F (2013) Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuel 6(5):89–89
Destruelle M, Holzer H, Klionsky DJ (1994) Identification and characterization of a novel yeast gene: the YGP1 gene product is a highly glycosylated secreted protein that is synthesized in response to nutrient limitation. Mol Cell Biol 14(4):2740–2754
Diao L, Liu Y, Qian F, Yang J, Yu J, Sheng Y (2013) Construction of fast xylose-fermenting yeast based on industrial ethanol-producing diploid Saccharomyces cerevisiae by rational design and adaptive evolution. BMC Biotechnol 13(1):1–9
Dimmer KS, Papić D, Schumann B, Sperl D, Krumpe K, Walther DM, Rapaport D (2012) A crucial role for Mim2 in the biogenesis of mitochondrial outer membrane proteins. J Cell Sci 125(Pt 14):3464–3473
Feng X, Zhao H (2013a) Investigating host dependence of xylose utilization in recombinant Saccharomyces cerevisiae strains using RNA-seq analysis. Biotechnol Biofuels 6(1):96
Feng X, Zhao H (2013b) Investigating glucose and xylose metabolism in Saccharomyces cerevisiae and Scheffersomyces stipitis via 13C metabolic flux analysis. AICHE J 59(9):3195–3202
Ferreira C, Lucas C (2007) Glucose repression over Saccharomyces cerevisiae glycerol/H+ symporter gene STL1 is overcome by high temperature. FEBS Lett 581(9):1923–1927
Flagfeldt DB, Siewers V, Huang L, Nielsen J (2009) Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast 26(10):545–551
Fu J, Hou J, Liu L, Chen L, Wang M, Shen Y, Zhang Z, Bao X (2013) Interplay between BDF1 and BDF2 and their roles in regulating the yeast salt stress response. FEBS J 280(9):1991–2001
Happel AM, Swanson MS, Winston F (1991) The SNF2, SNF5 and SNF6 genes are required for ty transcription in Saccharomyces cerevisiae. Genetics 128(1):69–77
Hamacher T, Becker J, Gardonyi M, Hahn-Hägerdal B, Boles E (2002) Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiol-SGM 148:2783–2788
Hector RE, Dien BS, Cotta MA, Mertens JA (2013) Growth and fermentation of D-xylose by Saccharomyces cerevisiae expressing a novel D-xylose isomerase originating from the bacterium Prevotella ruminicola TC2-24. Biotechnol Biofuels 6(1):1–12
Jaspersen SL, Giddings TH, Mark W (2002) Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J Cell Biol 159(6):945–956
Kim EM, Jang YK, Park SD (2002) Phosphorylation of Rph1, a damage-responsive repressor of PHR1 in Saccharomyces cerevisiae, is dependent upon Rad53 kinase. Nucleic Acids Res 30(3):643–648
Kim SR, Park YC, Jin YS, Seo JH (2013) Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism. Biotechnol Adv 31(6):851–861
Ko JK, Um Y, Woo HM, Kim KH, Lee SM (2016) Ethanol production from lignocellulosic hydrolysates using engineered Saccharomyces cerevisiae harboring xylose isomerase-based pathway. Bioresour Technol 209:290–296
Kratzer S, Schüller HJ (1995) Carbon source-dependent regulation of the acetyl-coenzyme a synthetase-encoding gene ACS1 from Saccharomyces cerevisiae. Gene 161(1):75–79
Larsson C, Påhlman IL, Gustafsson L (2000) The importance of ATP as a regulator of glycolytic flux in Saccharomyces cerevisiae. Yeast 16(9):797–809
Law DT, Segall J (1988) The SPS100 gene of Saccharomyces cerevisiae is activated late in the sporulation process and contributes to spore wall maturation. Mol Cell Biol 8(2):912–922
Lee SM, Jellison T, Alper HS (2014) Systematic and evolutionary engineering of a xylose isomerase-based pathway in Saccharomyces cerevisiae for efficient conversion yields. Biotechnol Biofuels 7(1):122
Li YC, Gou ZX, Liu ZS, Tang YQ, Akamatsu T, Kida K (2014) Synergistic effects of TAL1 over-expression and PHO13 deletion on the weak acid inhibition of xylose fermentation by industrial Saccharomyces cerevisiae strain. Biotechnol Lett 36(10):2011–2021
Li YC, Mitsumasu K, Gou ZX, Min G, Tang YQ, Li GY, Wu XL, Akamatsu T, Taguchi H, Kida K (2016a) Xylose fermentation efficiency and inhibitor tolerance of the recombinant industrial Saccharomyces cerevisiae strain NAPX37. Appl Microbiol Biotechnol 100(3):1531–1542
Li YC, Li GY, Gou M, Xia ZY, Tang YQ, Kida K (2016b) Functional expression of xylose isomerase in flocculating industrial Saccharomyces cerevisiae strain for bioethanol production. J Biosci Bioeng 121(6):685–691
Mabee W, Saddler J (2010) Bioethanol from lignocellulosics: status and perspectives in Canada. Bioresour Technol 101(13):4806–4813
Macías S, Bragulat M, Tardiff DF, Vilardell J (2008) L30 binds the nascent RPL30 transcript to repress U2 snRNP recruitment. Mol Cell 30(6):732–742
Madhavan A, Tamalampudi S, Ushida K, Kanai D, Katahira S, Srivastava A, Fukuda H, Bisaria VS, Kondo A (2009) Xylose isomerase from polycentric fungus Orpinomyces: gene sequencing, cloning, and expression in Saccharomyces cerevisiae for bioconversion of xylose to ethanol. Appl Microbiol Biotechnol 82(6):1067–1078
Mamnun YM, Schüller C, Kuchler K (2004) Expression regulation of the yeast PDR5 ATP-binding cassette (ABC) transporter suggests a role in cellular detoxification during the exponential growth phase. FEBS Lett 559(1–3):111–117
Matsushika A, Inoue H, Kodaki T, Sawayama S (2009) Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl Microbiol Biotechnol 84(1):37–53
Nakayama H, Yoshida K, Shinmyo A (2004) Yeast plasma membrane Ena1p ATPase alters alkali-cation homeostasis and confers increased salt tolerance in tobacco cultured cells. Biotechnol Bioeng 85(7):776–789
Palmqvist E, Hahn-Hägerdal B (2000) Fermentation of lignocellulosic hydrolysates. II: inhibitors and mechanisms of inhibition. Bioresour Technol 74(1):25–33
Park JS, Halegoua S, Kishida S, Neiman AM (2015) A conserved function in phosphatidylinositol metabolism for mammalian Vps13 family proteins. PLoS One 10(4):e0124836
Platara M, Ruiz A, Serrano R, Palomino A, Moreno F, Ariño J (2006) The transcriptional response of the yeast Na+-ATPase ENA1 gene to alkaline stress involves three main signaling pathways. J Biol Chem 281(48):36632–36642
Qi X, Zha J, Liu GG, Zhang W, Li BZ, Yuan YJ (2015) Heterologous xylose isomerase pathway and evolutionary engineering improve xylose utilization in Saccharomyces cerevisiae. Front Microbiol 6:1165
Rossouw D, Heyns EH, Setati ME, Bosch S, Bauer FF (2013) Adjustment of trehalose metabolism in wine Saccharomyces cerevisiae strains to modify ethanol yields. Appl Environ Microbiol 79(17):5197–5207
Shannon KW, Rabinowitz JC (1988) Isolation and characterization of the Saccharomyces cerevisiae MIS1 gene encoding mitochondrial C1-tetrahydrofolate synthase. J Biol Chem 263(16):7717–7725
Smith J, Van Rensburg E, Görgens JF (2014) Simultaneously improving xylose fermentation and tolerance to lignocellulosic inhibitors through evolutionary engineering of recombinant Saccharomyces cerevisiae harbouring xylose isomerase. BMC Biotechnol 14(1):1–17
Tang YQ, An M, Liu K, Nagai S, Shigematsu T, Morimura S, Kida K (2006) Ethanol production from acid hydrolysate of wood biomass using the flocculating yeast Saccharomyces cerevisiae strain KF-7. Process Biochem 41(4):909–914
Tanino T, Hotta A, Ito T, Ishii J, Yamada R, Hasunuma T, Ogino C, Ohmura N, Ohshima T, Kondo A (2010) Construction of a xylose-metabolizing yeast by genome integration of xylose isomerase gene and investigation of the effect of xylitol on fermentation. Appl Microbiol Biotechnol 88(5):1215–1221
Teste MA, Enjalbert B, Parrou JL, François JM (2000) The Saccharomyces cerevisiae YPR184w gene encodes the glycogen debranching enzyme. FEMS Microbiol Lett 193(1):105–110
Tibbetts AS, Sun Y, Lyon NA, Ghrist AC, Trotter PJ (2002) Yeast mitochondrial oxodicarboxylate transporters are important for growth on oleic acid. Arch Biochem Biophys 406(1):96–104
Tolliday N, Pitcher M, Li R (2003) Direct evidence for a critical role of myosin II in budding yeast cytokinesis and the evolvability of new cytokinetic mechanisms in the absence of myosin II. Mol Biol Cell 14(2):798–809
van Rensburg E, den Haan R, Smith J, van Zyl WH, Görgens JF (2012) The metabolic burden of cellulase expression by recombinant Saccharomyces cerevisiae Y294 in aerobic batch culture. Appl Microbiol Biotechnol 96(1):197–209
Vilela LD, de Araujo VPG, Paredes RD, Bon EPD, Torres FAG, Neves BC, Eleutherio ECA (2015) Enhanced xylose fermentation and ethanol production by engineered Saccharomyces cerevisiae strain. AMB Express 5(1):16
Wang G, Tan L, Sun ZY, Gou ZX, Tang YQ, Kida K (2015) Production of bioethanol from rice straw by simultaneous saccharification and fermentation of whole pretreated slurry using Saccharomyces cerevisiae KF-7. Environ Prog Sustain Energy 34(2):582–588
Zeng W-Y, Tang Y-Q, Gou M, Xia Z-Y, Kida K (2016) Transcriptomes of a xylose-utilizing industrial flocculating Saccharomyces cerevisiae strain cultured in media containing different sugar sources. AMB Express 6:51
Zhang Z, Moo-Young M, Chisti Y (1996) Plasmid stability in recombinant Saccharomyces cerevisiae. Biotechnol Adv 14(4):401–435
Zhao Q, Pan L, Ren Q, Hu D (2015) Digital gene expression analysis in hemocytes of the white shrimp Litopenaeus vannamei in response to low salinity stress. Fish Shellfish Immun 42(2):400–407
Zhou H, Cheng JS, Wang BL, Fink GR, Stephanopoulos G (2012) Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab Eng 14(6):611–622
Zhou YJ, Buijs NA, Siewers V, Nielsen J (2014) Fatty acid-derived biofuels and chemicals production in Saccharomyces cerevisiae. Front Bioeng Biotechnol 2:32
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31170093) and the Talent Project for Science and Technology Innovation of Sichuan Province (2017RZ0021).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared no conflict of interest, and this article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 1328 kb).
Rights and permissions
About this article
Cite this article
Li, YC., Zeng, WY., Gou, M. et al. Transcriptome changes in adaptive evolution of xylose-fermenting industrial Saccharomyces cerevisiae strains with δ-integration of different xylA genes. Appl Microbiol Biotechnol 101, 7741–7753 (2017). https://doi.org/10.1007/s00253-017-8494-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8494-z