Abstract
With the increased knowledge on spore structure and advances in biotechnology engineering, the newly developed spore-surface display system confers several inherent advantages over other microbial cell-surface display systems including enhanced stability and high safety. Bacillus subtilis is the most commonly used Bacillus species for spore-surface display. The expression of heterologous antigen or protein on the surface of B. subtilis spores has now been practiced for over a decade with noteworthy success. As an update and supplement to other previous reviews, we comprehensively summarize recent studies in the B. subtilis spore-surface display technique. We focus on its benefits as well as the critical factors affecting its display efficiency and offer suggestions for the future success of this field.
Similar content being viewed by others
Introduction
Spore-surface display, which is a powerful technology that can effectively express bioactive molecules on the surface of spores, is becoming increasingly important. In the development of spore-surface display, Bacillus subtilis is recognized as the most commonly used Bacillus species because of the following: (1) recent advances in knowledge of its spore structure (Driks 1999; Henriques and Moran 2007), (2) amenability and ease of well-established genetic manipulation (Cutting and Vander-Horn 1990), and (3) genomic data (Kunst et al. 1997). Owing to enhanced stability and high safety (Hong et al. 2008; Cutting 2011), the emergence of new recently developed B. subtilis spore-surface display that enables the display of various food- or human-related antigens or enzymes on the spore surface for the development of vaccines and whole-cell biocatalysts has helped to advance the field of display methodology (Isticato et al. 2001; Sibley et al. 2014; Kwon et al. 2007; Xu et al. 2011a).
B. subtilis spore display, which possesses exogenous antigen or protein fused to one of the spore coat proteins on the spore surface, celebrates its 15th anniversary in 2016. The initial concept was proposed in 2001 by Isticato and coworkers. Since then, the use of this technology has steadily been progressing due to innovations introduced and more anchoring motifs available. Several excellent recent reviews of independent topics described Bacillus spore display (Lee et al. 2003; Wu et al. 2008; Kim and Schumann 2009; Knecht et al. 2011; Pan et al. 2012; Isticato and Ricca 2014), but a comprehensive review of B. subtilis spore display is rare. In this review, we highlight new developments in the field of B. subtilis spore-surface display. In addition, the prospects of future spore-surface display development are briefly discussed.
Composition of B. subtilis spore
In the Bacillus genus, the Gram-positive bacterium B. subtilis is one of the most widely used species for the production of extracellular enzymes on an industrial scale (Schallmey et al. 2004). When confronted by nutrient depletion, the normally rod-shaped B. subtilis cells produce a morphologically distinct cell type called the spore. A typical fine structure of the B. subtilis spore is shown in Fig. 1. The spore consists of a central core compartment that contains a copy of the chromosome and several layers including proteinaceous spore coat, outer forespore membrane, cortex, and inner forespore membrane. The multilayered spore coat protects the spore against bacterial enzymes, toxic chemicals, and harsh environment. In B. subtilis, the coat is made up of a minimum of 70 proteins unique to spores, which comprises four distinct layers: basement layer, inner coat, outer coat, and crust (McKenney et al. 2013) (Fig. 1). The outer forespore membrane, located between coat and cortex, also plays an important role in spore formation (Piggot and Hilbert 2004). The cortex, a thick layer of peptidoglycan, is responsible for maintaining the highly dehydrated state of the core, thereby contributing to the development and maintenance of spore resistance (Imamura et al. 2011). The inner forespore membrane plays a major role in spore resistance to numerous chemicals (Setlow 2006).
Overview of B. subtilis spore display
B. subtilis is classified as generally recognized as safe (GRAS) status by the US Food and Drug Administration, and it has been proposed as a probiotic in both human and animal uses (Huang et al. 2008; Hong et al. 2006; Cutting et al. 2009). In addition, B. subtilis spores can be easily engineered to display heterologous antigen or protein on their surface. However, such genetically modified B. subtilis spores do not affect spore structure and their spore resistant properties under extreme conditions (Hinc et al. 2013). Other advantages include the facts that the fusion protein has not to pass through any membrane barrier and there is no bias toward codon usage. These attributes make B. subtilis spore a novel and interesting platform for the display of bioactive molecules. B. subtilis spore display is accomplished by inserting a foreign gene encoding an antigen or protein into a B. subtilis gene encoding a component of the spore coat. The ideal spore coat protein should only act as an anchor for the displayed antigen and do not interfere with its structure. Moreover, the insertion site of the fusion between the target and anchor protein can affect its stability, activity, and post-translational modification to a certain extent. For minimizing this steric hindrance effect, N-terminal, C-terminal, and sandwich fusions have been introduced. Notably, spore display can provide the target antigen or protein free access to its soluble substrate.
Based on the mechanism of sporulation, a schematic illustrating a possible process for the development of spore-surface display is presented in Fig. 2. The initial step is to transform the recombinant plasmid containing a spore coat protein and heterologous protein fusion gene into the competent B. subtilis cells. Then, actively growing cells of recombinant B. subtilis are induced to differentiate into spores. As sporulation proceeds, the rod-shaped cell undergoes an unequal cell division into two compartments, namely, a larger mother cell and a smaller forespore. Subsequently, the cot genes encoding spore coat proteins are expressed within the mother cell compartment. Once engulfment is completed, the coat proteins and fusion with coat proteins produced inside the mother cell assemble on the forespore. As a consequence, the forespore is surrounded by two membranes. Eventually, cell lysis occurs in a programmed event to release the mature spore displaying the heterologous protein on its surface into the environment.
Until recently, various microbial cell-surface display systems, such as phage display, Escherichia coli cell display, and yeast cell display, have been developed. The advantages and disadvantages of B. subtilis spore display and some of the extensive studied surface display systems are illustrated in Table 1. Phage display is the first successful approach for displaying foreign polypeptide on the surface of the phage particle (Smith 1985) and has since been applied for biotechnological and biomedical fields. However, the use of phage display is inherently restricted by the simple post-translation modification and the presentation of smaller functional proteins. Similar to phage display, E. coli and yeast cell display systems present their own shortages. The primary disadvantage of the former technology is its low safety, whereas relatively low stability and prolonged fermentation period are the main drawbacks of the latter. For bypassing several problems associated with traditional display systems, B. subtilis spore display has been established. Compared with other display technologies, this technique offers additional advantages, such as higher safety and enhanced stability, suggesting that B. subtilis spore display is a good molecular display platform to surface express food/human-related proteins or thermolabile enzymes. With the spore-based display system, large and complex enzymes have been expressed on the spore surface. Therefore, B. subtilis spore display is a powerful alternative to traditional display systems.
Factors affecting the efficiency of B. subtilis spore display
A “one size fits all” surface display system does not exist. The principle and procedure of B. subtilis spore display is simple and easy to perform, but the outcome of display efficiency can vary depending upon two elements. One is the amount of expressed protein on the spore surface, and the other is the activity and stability of the displayed protein. More generally, numerous factors affect the efficiency of B. subtilis spore display system, including the localization and properties of the anchor protein, the nature of the displayed target protein, how this is linked to the spore coat protein, the type of expression vector and host strain, the use of a flexible peptide linker between the spore coat protein and the target protein, and other multiple experimental parameters.
Selection of an appropriate anchor protein is a critical factor in the successful presentation of foreign antigen or protein by B. subtilis spore display. The localization and characteristics of the coat protein apparently acts as a vital parameter in candidate anchor protein ranking. Discordance of display efficiency was observed even for the same target protein by using different coat proteins as anchoring motifs. Examples of these proteins are urease subunit A (UreA), flagellar cap protein, α-amylase, GFPuv, and β-galactosidase (Hinc et al. 2010b; Negri et al. 2013; Nguyen and Schumann 2014; Wang et al. 2015, 2016). The size and properties of the displayed target protein are key factors in determining B. subtilis spore display efficiency. However, this display technology may tend to fail when applied to the larger and more complex target proteins. Utilizing a high copy number of E. coli–B. subtilis shuttle vector, especially non-integrative recombinant plasmid, with the appropriate promoter was shown to exert a beneficial effect on the amount of displayed protein (Xu et al. 2011a; Nguyen and Schumann 2014; Iwanicki et al. 2014; Chen et al. 2015a). In addition, the selection of the appropriate flexible linker inserted into the correct fusion between the anchor protein and the displayed target protein may be essential for stable expression in certain cases (Hinc et al. 2013). Given that the target protein is located on the spore surface surrounded by other coat proteins, the surface expression is partially influenced by several abundant and non-fully cooperative spore coat proteins, resulting in lower display efficiency. Deletion of genes encoding the non-fully cooperative coat proteins need to be taken into consideration. Besides the abovementioned factors, the selection of an appropriate host strain is another important factor that affects the display performance of the targeted protein. Cultivation conditions also exert an important influence on the activity and/or the amount of displayed target protein. In general, the display efficiency is extremely challenging to improve, and a careful assessment of numerous factors is required to explore the possible factors that strongly affect display efficiency.
Conventional B. subtilis spore display system
The first report on spore-surface display dates from 2001 (Isticato et al. 2001). In the subsequent years, a variety of other spore coat proteins, such as CotC, CotG, CotE, CotX, CotZ, CgeA, and OxdD, have been identified as suitable anchoring motifs that display heterologous antigen or protein on the spore surface of B. subtilis. A summary of the published studies on the B. subtilis spore-surface display is given in Table 2. Over the past 15 years, when CotB was used as an anchor motif, DNA encoding the three 27 amino acid (aa) repeats located at the C-terminus of CotB was typically omitted, and only DNA encoding the N-terminal 275 aa residues of CotB was used possibly because three repeat fragments cause genetic instability on chimeric proteins containing them (Isticato et al. 2001). In certain cases, however, the full length version of CotB has also been fused to express Bacillus anthracis and Clostridium perfringens antigens (Duc et al. 2007; Hoang et al. 2008). When all other anchor proteins except CotB were used as fusion partners, DNA encoding the entire Cot proteins was applied. For obtaining correctly expressed fusion protein, the natural promoter of the cot gene is routinely used to control the proper timing of expression of the foreign gene during the sporulation process. The majority of spore display examples often rely on stable integration of an expression construct into the chromosome of B. subtilis. However, this approach presents several limitations, such as low expression level and complex operation. Utilizing a high copy number E. coli–B. subtilis shuttle vector can help to overcome these problems. As shown in Table 2, CotB and CotC have been successfully used to anchor different antigens on the spore surface for vaccine development, whereas CotG was frequently applied to target functional enzyme display onto the spore for whole-cell catalyst preparation. Until recently, effective antigen presentation by either CotZ or CgeA for identifying potential vaccine candidate has also been reported. CotC has sometimes appeared to be a good candidate for the display of alcohol dehydrogenase (Wang et al. 2011; Yuan et al. 2014) and β-galactosidase (Tavassoli et al. 2013). However, in addition to CotG and CotC, OxdD, CotX, and CotE served as anchoring motifs for proteins such as β-glucuronidase (Potot et al. 2010), L-arabinose isomerase (Liu et al. 2014), or tyrosinase (Hosseini-Abari et al. 2016).
Concerning the conventional B. subtilis spore display, the analysis of this approach will be discussed in two aspects according to the number of anchor proteins used.
B. subtilis spore display using an individual spore coat protein as an anchoring motif
CotB
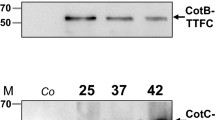
CotB is a 46-kDa protein located in outer layer of B. subtilis spore coat. The first evidence that a model antigen C-terminal fragment of tetanus toxin (TTFC) (Medaglini et al. 2001) can be expressed on the surface of B. subtilis PY79 spores by fusion of it to CotB was described by Isticato et al. (2001). In that study, only spores expressing the C-terminal CotB-based fusion protein were able to provide some protection against tetanus in mice. Later on, Duc et al. (2003b) studied mice orally or intranasally immunized with CotB–TTFC. They evaluated the effects on the biological activity (i.e., elicited antitoxin response and the protective efficacy) of these mice, which displayed B. subtilis spores. Unexpectedly, the spore-displayed TTFC can survive transit in the digestive tract. Moreover, a significant protection against a lethal dose of tetanus toxin is also detected in mice orally immunized with recombinant spores.
In 2004, the use of a combined vaccination strategy termed mucosal priming–parenteral boost route to generate strong immune responses against tetanus toxin challenge was explored (Ciabattini et al. 2004). This immunization schedule resulted in detectable levels of TTFC-specific IgA and IgG responses in both BALB/c and C57BL/6 mice when orally primed with recombinant spores presenting TTFC. Uyen et al. (2007) found that responses against Clostridium tetani TTFC (CsTTFC) were enhanced by the use of a combined expression of CsTTFC with spore display and germinating spores. In addition, CsTTFC expressed within the germinating spore provided significantly greater protection against tetanus compared with spore-displayed CsTTFC. Another study revealed that BALB/c mice orally immunized with recombinant spores expressing TTFC on the surface of both germination-defective and congenic wild-type strains stimulated a strong cellular immune response with a Th1 phenotype (Mauriello et al. 2007). In this case, this immune response is independent of spore germination in the animal gastrointestinal tract (GIT). In addition to TTFC, the CotB-mediated spore display was investigated to target other antigens for developing candidate vaccines against necrotic enteritis, enterovirus 71, white spot syndrome virus (WSSV), H1N1 influenza virus, and tuberculosis (Table 2). In these cases, the resulting vaccines were able to elicit antigen-specific immune responses and are protective in animals. All these data clearly demonstrated the use of recombinant spores as a good vaccine vehicle.
In principle, the B. subtilis spore-based vaccine can be divided into two classes, namely, recombinant vaccines (spore expressing antigen on its surface) and non-recombinant vaccines (spore-based adsorption of antigen). In both cases, mice immunized with either the recombinant spores or adsorbed spores were demonstrated to be effective and safe in providing protection against infection (Duc and Cutting 2003a; Oggioni et al. 2003; Huang et al. 2010; Permpoonpattana et al. 2011). Sibley et al. (2014) have recently highlighted that both live and formaldehyde-inactivated recombinant spores surface-expressing the MPT64 antigen could elicit Th1 responses in mice, thus proving the effectiveness of the developed TB vaccine against tuberculosis challenge in the murine model. Most importantly, the use of such inactive recombinant spores exerts no ethical consideration associated with the deliberate release of GMOs (Sibley et al. 2014).
Given the extremely short residence time of a recombinant spore-based vaccine in mouse GIT, the oral administration of this type of vaccine is often prone to evoking poor immune responses (Unnikrishnan et al. 2012). This inherent limitation may be overcome by using “gut-colonizing”, a modified B. subtilis 1012 spore-based, mucosal vaccine delivery system. This system consists of two antigen expression strategies (Batista et al. 2014). The first one contains two different bacterial adhesins, S-layer protein (SlpA) from Lactobacillus brevis ATCC 8287 and invasin (InvA) from Yersinia pseudotuberculosis. The second one involves P1 protein of Streptococcus mutans NG8 as a model antigen. Apart from the presentation of P1 on the spore surface mediated by CotB, one bacterial adhesin was also expressed in germinated spores. Through this strategy, the expressed bacterial adhesin in the germinated spores facilitates adhesion onto the mucosal surface of animal gut, thereby enabling CotB-P1-displaying recombinant spores to prolong transit through the GIT. Mice immunized by sublingual administration of three doses of B. subtilis spores produced higher titers of specific IgG in their serum than mice vaccinated with a ninefold higher dose of spores of the same strains via oral vaccination route (Batista et al. 2014).
Besides its well-known reputation in the development of vaccine vehicles, the CotB-mediated B. subtilis spore display has been explored as a potential platform for drug delivery. In cancer therapy, current targeting delivery of anti-cancer drugs to cancerous cells suffers from several drawbacks. The concept of using B. subtilis spore display as anti-cancer drug delivery agent was initially developed by Nguyen et al. (2013). For achieving this goal, a clever strategy that relies on a combination of two approaches was applied. One approach is based on the expression of streptavidin as a fusion to the CotB protein at the spore coat, and a second approach involves the adsorption of diterpen paclitaxel, a mitotic inhibitor used for cancer treatment, to the CotB-streptavidin-displaying recombinant spores. By this strategy, the obtained recombinant spores termed “paclitaxel–spore–cetuximab” biocomplex could recognize a primary biotinylated antibody, followed by selective binding to the human epidermal growth factor receptor (EGFR) on HT 29 colon cancer cells, thus enabling the target anti-cancer drug to efficiently suppress cancerous cell division and proliferation (Nguyen et al. 2013). The findings of this study highlight that these recombinant spores displaying streptavidin appear to be an attractive drug delivery system by conjugating with an appropriate biotinylated antibody.
Using CotB as a fusion partner, Chen et al. (2015a) obtained a spore display of a thermophilic, alkali tolerant, and methanol-activated lipase Tm1350 produced by Thermotoga maritima MSB8 (ATCC 43589). After spore display, the optimal temperature increased by 10 °C (from 70 to 80 °C) and its optimal pH shifted from 7.5 to 9. Furthermore, the spore-displayed Tm1350 was found to exhibit an increase in thermal stability, pH stability, and reusability compared with those of its free form. In another example, the successful display of a novel thermostable esterase from Clostridium thermocellum at the B. subtilis spores was achieved with the same anchor protein (Chen et al. 2015b). The resulting spore-displayed esterase not only exhibited a broad temperature and pH optimum but also showed enhanced tolerance to DMSO. The evident improvement in stability of spore-displayed enzymes was probably due to the introduction of a flexible linker (GGGGS) and the use of extrachromosomal expression system from a high copy number plasmid. The overall conclusion of both studies was that the CotB-mediated B. subtilis spore display may be a useful tool for enzyme immobilization.
As the above examples demonstrate CotB-mediated spore display for development as vaccine and whole-cell biocatalyst, this approach could also be applied as a bioremediation tool (Hinc et al. 2010a). Spores of B. subtilis strain engineered to effectively express 18 histidine residues were demonstrated to be an efficient bioadsorbent for nickel removal. In addition, the efficiency of nickel binding is not influenced by either pH or temperature but is strongly dependent on the amount of spores used in the adsorption reaction (Hinc et al. 2010a).
CotC
CotC (12-kDa) is another spore coat protein in the B. subtilis. TTFC has also been displayed as a fusion to CotC as well as the heat-labile enterotoxin B subunit of E. coli (LTB); both antigen-displaying spores generated a positive immune response in mice (Mauriello et al. 2004). To improve the display efficiency of the CotC–TTFC fusion protein, both N- and C-terminal fusions were investigated by Isticato et al. in 2007. A fivefold increase in display efficiency was achieved when TTFC was expressed at the N-terminal end of CotC. The effectiveness of the N-terminal fusion approach has also been demonstrated with two T helper cell epitopes pep23 (residues 249–263 of HIV-1 reverse transcriptase) and pep24 (residues 191–205 of HIV-1 gp120) (D’Apice et al. 2007).
Considering both the safety and low-cost production, B. subtilis spore display presents a valuable application in vaccine design and delivery. As summarized in Table 2, CotC is mainly used as an anchoring motif for vaccine development. Zhou et al. (2008a, 2008b) searched for an alternative vaccine against clonorchiasis, a disease caused by infection with Clonorchis sinensis. They found two putative tegumental proteins (TP20.8 and TP22.3 of C. sinensis) that can elicit a significant level of protection in vaccinated rats when delivered as an oral recombinant spore-based vaccine. In recent studies, other enzymes reported to be alternative candidates for the use in vaccine development against C. sinensis prevention are enolase of C. sinensis (Csenolase) and leucine aminopeptidase 2 of C. sinensis (CsLAP2) (Wang et al. 2014; Qu et al. 2014a). The study of Yu et al. (2015) further evidenced that oral immunization in rats with recombinant spores displaying Csenolase induces both systemic and local mucosal immune responses. In addition, other vaccine antigens, including glutathione S-transferase of Schistosoma japonicum, envelope fusion glycoprotein GP64 of Bombyx mori nucleopolyhedrovirus (BmNPV), urease B protein of Helicobacter pylori (HpUreB), and VP26 of WSSV were also fused to the C-terminal end of CotC. The immunogenicity of the resulting recombinant spores has been assessed in mouse models in some cases with highly promising results. The reports supporting the use of the CotC-mediated spore display as an ideal vaccine delivery system will continue to grow.
Current interest is focused in utilizing B. subtilis spore display as a new delivery vector of therapeutic protein. To date, human serum albumin (HSA), human proinsulin (HPI), and human growth hormone (HGH) have been displayed on the spore surface by using CotC as an anchoring motif. In these cases, the CotC-linked fusion genes were stably integrated into the B. subtilis chromosome. The in vivo study demonstrated that mice immunized orally with HSA-expressing spores are capable of increasing blood serum albumin level in a dose-dependent manner (Mao et al. 2012). Although HSA fused to CotC is less efficiently displayed, this spore display format offers several advantages, including high stability and easy production over the yeast cell expression system (Mao et al. 2012). In an alternative approach to the use of mammals for drug development, the same research group studied the application of silkworm for evaluating the therapeutic effects of the recombinant spores expressing HPI or HGH (Feng et al. 2013; Lian et al. 2014). Both studies confirm the efficacy of the recombinant spores that can be digested and absorbed into silkworm hemolymph. The authors reported that this result can presumably be explained by multiple mechanisms, such as the addition of the enterokinase site, inherent resistant properties of B. subtilis 168 spores, or the simple intestinal structure of silkworm.
Whereas most of the displayed enzymes tend to be accomplished by fusion to CotG, several recent studies demonstrated that heterologous enzymes on the surface of B. subtilis spores can be targeted through the CotC anchoring motif. The first enzyme displayed on B. subtilis 168 spores via fusion with CotC was alcohol dehydrogenase from B. mori (BmADH) (Wang et al. 2011). Compared with the native enzyme, the spore-displayed BmADH exhibits higher enzymatic activity and stability. The β-galactosidase of B. subtilis 168 (LacA), a rather large protein of 116 kDa being active as a homotetramer, is another example (Tavassoli et al. 2013). In this study, the gene fusion of cotC–lacA was chromosomally integrated into B. subtilis 168 and a mutant B. subtilis RH101 (CotC-deficient strain), respectively. Western blotting confirmed the expression of the fused cotC–lacA at the spore surface of both 168 and RH101 strains. Indeed, both of spore-displayed LacAs could still retain relatively high activities even after exposure to different harsh environmental conditions. An interesting observation in the case of the strain deleted for cotC gene is that the CotC–LacA showed better reusability compared with that surface-expressed by B. subtilis 168, indicating a higher thermal stability for the spore-displayed LacA. The observed difference may be attributed to the difference in genetic background between 168 and RH101 strains (Tavassoli et al. 2013). A third example is to target alcohol dehydrogenase A from Acetobacter pasteurianus ET-7-3 at the spore surface by fusing it with CotC, yielding a modified B. subtilis 168 strain with enhanced ethanol tolerance compared with wild-type spores, which offers an attractive alternative for the liquor industry (Yuan et al. 2014).
CotG
CotG is a 24-kDa protein produced in the outer layer of B. subtilis spore coat. Kim et al. (2005) fused the biotin-binding streptavidin to the CotG protein of B. subtilis DB104. This protein is the first example of a tetrameric protein anchored on the spore surface. Surface-displayed streptavidin was verified by fluorescence-activated cell sorting and immunological methods. As demonstrated with the CotB-fused anti-cancer drug delivery agent (Nguyen et al. 2013), spore-displayed streptavidin can be a widely used tool in the detection of biotin-conjugated bioactive molecules. GFPuv, one of the most important engineered variants of green fluorescent protein (GFP) (Crameri et al. 1996), along with fusion to CotG, has become an efficient tool for the rapid identification of other possible spore coat proteins in B. subtilis (Kim et al. 2007). Another coat protein, CotX, has been proven useful for developing B. subtilis spore display and is the result of using this GFPuv as a reporter protein (Li et al. 2011). In a recent work (Mou et al. 2016), transmissible gastroenteritis virus spike (TGEV-S) protein is anchored onto the spores of B. subtilis WB800. Following the vaccination of suckling piglets with this recombinant spores, elevated specific SIgA titers in feces, IgG titers, and neutralizing antibodies in serum are generated in response to the in vivo stimulation with TGEV-S antigen. Results show that following TGEV challenge piglets that are passively immunized with the recombinant spores presenting the TGEV-S become highly protected against TGEV via dendritic cells. These findings serve as a valuable basis for developing an appropriate TGEV vaccine by using this spike protein.
For obtaining an enzyme with desirable properties, CotG is often applied as an anchoring motif for the surface expression of native enzymes. The β-galactosidase of E. coli (Ecβ–Gal), the first enzyme to be displayed with CotG, is only stable under mild conditions (Kwon et al. 2007). However, the spore-displayed Ecβ–Gal not only exhibits an increased tolerance to non-polar hydrophobic organic solvents but could further be stabilized via cross-linking with glutaraldehyde. Furthermore, this spore-displayed Ecβ–Gal can serve as a new type of biocatalyst to efficiently synthesize octyl-β-D-galactopyranoside from lactose and octanol (Kwon et al. 2007). In addition to Ecβ–Gal, other enzymes such as N-acetyl-D-neuraminic acid (Neu5Ac), ω-transaminase of Vibrio fluvialis JS17, meta-cleavage product hydrolase (MfphA and BphD) of Dyella ginsengisoli LA-4, and nitrilases of both T. maritima MSB8 (ATCC 43589) and C. thermocellum were also fused to the C-terminus of CotG. According to the results of Xu et al. (2011a), the spore-displayed Neu5Ac aldolase produces a high concentration of Neu5Ac (54.7 g/L) with a yield of 90.2% under optimal conditions. Compared with this new synthetic strategy, virtually the same concentration of Neu5Ac (53.9 g/L) but a lower yield (17.4%) was achieved by a coupled chemoenzymatic process (Gao et al. 2011). Considering the advantages of good stability and easy purification over free or immobilized enzyme, the use of such a spore-displayed Neu5Ac aldolase offers promise for Neu5Ac production. Hwang et al. (2011) reported that after spore display, V. fluvialis JS17 ω-transaminase, a cofactor-containing enzyme, exhibited a 30-fold higher enzymatic activity compared with wild-type spores. Another recent research of Qu et al. (2014b) showed that MfphA and BphD from D. ginsengisoli LA-4 show increased thermal and pH stabilities as a result of their surface display on spores using CotG as an anchoring motif. Moreover, this spore display not only facilitates the recovery of MfphA and BphD from the reaction mixture but still retains 45 and 70% of their initial activities even after ten consecutive runs, respectively. One of the recent applications of the CotG protein is in the spore display of nitrilase from T. maritima MSB8 (ATCC 43589) (Chen et al. 2016). After spore display, the optimal temperature increased by 5 °C (from 45 to 50 °C) and its optimal pH shifted from 7.5 to 8. Besides, spore display induced an increase in thermal stability and pH stability compared with the free form. Furthermore, the spore-displayed nitrilase retained over 80% of initial activity after five successive reaction cycles. A similar result was obtained in case of nitrilase from C. thermocellum by the CotG-mediated spore display method (Chen et al. 2015c).
Altogether, the presented examples clearly indicate that among all anchoring motifs reported to date, the CotG-mediated spore display system is excellent for the development of whole-cell biocatalysts.
CotX
CotX has recently been identified as a crust protein (17 kDa) (Imamura et al. 2011). Previously, CotX has been exploited for surface display by fusion to the most utilized reporter protein GFP (Li et al. 2010). Although significant fluorescence could also be detected around the recombinant spores harboring the CotX-linked GFP fusion protein, several advanced analytical methods, such as western blotting and flow cytometry analyses, were not applied to confirm the presence of the protein. Similar to the case of Ecβ–Gal, no data was available for the verification of the surface expression of Ecβ–Gal on B. subtilis spores (Kim et al. 2004). However, recent studies begin to demonstrate the feasibility of the CotX-mediated spore display. In the study by Liu et al. (2014), L-arabinose isomerase (L-AI) from Lactobacillus fermentum CGMCC 2921 was successfully expressed on the surface of B. subtilis 168 spores through fusion with CotX. After being displayed, L-AI exhibits a relatively high activity and improved stability. In addition, a 1.4-fold increase in conversion rate of D-galactose was obtained with this spore-displayed L-AI compared with the previously known system, wherein the highest conversion rate was obtained with the free L-AI (Xu et al. 2011b; Liu et al. 2014). Further evidence of the applicability of the CotX-mediated spore display was found in an example from the β-galactosidase of Bacillus stearothermophilus IAM11001 (Bsβ-Gal) (Wang et al. 2016). After spore display, the optimum temperature shifted from 70 to 75 °C and its pH optimum decreased by 0.5 pH unit (to pH 6.0). Compared with the free Bsβ–Gal, spore display could also lead to high thermal tolerance and repeated reuse. In addition, with lactose and fructose as substrates, this spore-displayed Bsβ–Gal produced 8.8 g/L lactulose with a yield of 4.4% (Wang et al. 2016). The results of these two studies validate CotX-mediated spore display as an effective tool in the development of a novel biocatalyst with improved enzyme performance.
OxdD
OxdD is a 43-kDa minor component of the spore coat. Recently, a successfully heterologous surface expression was achieved in B. subtilis PY79 by fusion of a monomeric phytase, a commonly used feed enzyme that assists in the breakdown of undigestible phytic acid, to an inner coat protein OxdD (Potot et al. 2010). Spore-displayed phytase was found to increased the nutritional value of feed by liberating the phytase-bound phosphorous, thus lowering the demand for supplementation of inorganic phosphorous. The authors also compared OxdD with CotG to judge whether the former exhibits higher display efficiency for phytase and could be used more frequently for other proteins. As reported, the CotG–phytase fusion protein showed a twofold increase in enzymatic activity compared with fusion to OxdD. This finding led to the proposal that the location of OxdD within the coat may protect the enzyme from the environment but hinder the access of the substrate to the active site of the spore-displayed phytase. Conversely, protection of the displayed enzymes conferred by the OxdD-mediated spore display system may be advantageous in certain applications, especially in the production of orally ingested spore-based probiotic products, as their activities could be well maintained even through the highly acidic and proteolytic environment of the stomach. Moreover, the prospect of OxdD as an anchoring motif was further illustrated by the successful display of a bioactive tetrameric β-glucuronidase from E. coli K-12 on the spore surface (Potot et al. 2010).
CotZ
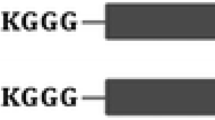
CotZ, a 16-kDa protein required for the assembly of the newly identified spore crust, has been found to act as a new anchoring motif for the efficient display of UreA of Helicobacter acinonychis on the spores (Imamura et al. 2011; Hinc et al. 2013). In the case of the CotZ–UreA fusion protein, the calculated number of recombinant protein molecules is 2.5 × 102 from a single spore. This fusion protein is more effective in stimulating immunological response than other antigens in mice. Hinc et al. (2013) attempted to gain insight into the effect of an additional peptide linker on the display efficiency of H. acinonychis UreA as a fusion with CotB. They correctly introduced two peptide linkers (GGGGS and GGGEAAAKGGG) between CotB and UreA. However, the CotB–GGGGS–UreA fusion protein failed to be displayed on the spore surface possibly because the inserted short linker cannot form any secondary structure and is thus unable to efficiently stabilize the structure. In contrast, experiments with a longer linker generated promising results, as further demonstrated by western blotting, immunofluorescence microscopy, and dot blot analyses. Furthermore, the expression level of the CotB–GGGEAAAKGGG–UreA fusion protein (1.0 × 104 recombinant molecules per spore) is remarkably higher than that of the CotB–UreA fusion protein without the insertion of an appropriate peptide linker (1.1 × 103) (Hinc et al. 2010b, 2013); this finding states that incorporation of this flexible linker between CotB and UreA resulted in improved display efficiency. Similar results were obtained with FliD when expressed in a same approach for CotB–GGGEAAAKGGG–UreA fusion protein (Negri et al. 2013). This interesting phenomenon may be attributed to the insertion of the flexible longer linker that could form a stable α-helical structure during post-translational processing of the fusion protein.
CotE
In addition to CotG and CotC, another outer coat protein CotE (21 kDa) was used as an anchoring motif for the successful immobilization of Ecβ–Gal at the surface of B. subtilis DB104 spores (Hwang et al. 2013). However, the incorporation of CotE–Ecβ–Gal fusion protein resulted in disruption of the intact structural integrity of recombinant spore wall, whereas the inserted Ecβ–Gal fused to CotG exerted no effect on its integrity. This observation provides certain support that the morphogenetic protein CotE is essential to maintain the integrity of the spore wall (Ozin et al. 2001; McKenney and Eichenberger 2012). In addition, the thermal stability of Ecβ–Gal remained unchanged either for free or for spore-displayed, which is in poor agreement with the previous report (Kwon et al. 2007). Different from CotE–Ecβ–Gal, CotE–Tyr (a successful spore display of Bacillus megaterium DSM319 tyrosinase based on the use of CotE) demonstrated good stability and reusability after spore display (Hosseini-Abari et al. 2016). The insertion of a suitable linker (GGGGS) between CotE and Tyr and the use of extrachromosomal expression system for Tyr may partially explain this unique catalytic behavior.
CgeA
Similar to CotZ, CgeA is another 14-kDa crust protein, and its possibility in the surface expression of H. pylori NCTC 11637 cytotoxic-associated gene A protein (CagA) on the B. subtilis 168 spores was investigated (Iwanicki et al. 2014). Both CgeA–CagA and CgeA–GGGEAAAKGGG–CagA fusion proteins were successfully displayed on the surface of B. subtilis 168 spores, as confirmed by western blotting and immunofluorescence microscopy analyses. Moreover, the recombinant spores expressing CgeA–linker–CagA exhibited stronger fluorescent signal than recombinant spores displaying CgeA–CagA. When these data are considered together with the data on UreA and FliD, the use of a flexible peptide linker (GGGEAAAKGGG) is suggested to considerably improve the display efficiency of the targeted protein. To further enhance the display efficiency, a universal vector was constructed by introducing an α-helical linker coupled with a selection marker trophic gene (Iwanicki et al. 2014). Given the lack of antibiotic resistance genes in this vector, this approach would offer improved safety in the development of spore-based vaccine or whole-cell biocatalyst (Iwanicki et al. 2014).
B. subtilis spore display using two or more independent coat proteins as anchoring motifs
The inherent benefits of the use of B. subtilis spores as antigen delivery platform offer an attractive second-generation vaccine vehicle (Amuguni and Tzipori 2012). When developing an oral vaccine against anthrax caused by the bacterium B. anthracis, protective antigen (PA) is often selected because of its stable and highly immunogenic properties. In the study of Duc et al. (2007), the target of PA fragment was displayed on the spore surface by fusing it with CotB or CotC. Full-length PA was also expressed in the germinated spores. Moreover, the highest levels of protective immunity in mice were observed when orally immunized only with recombinant strain, enabling the efficient production of both surface display of PA fragment on the spores and, simultaneously, the secretion of PA from germinating spores. However, the vaccine obtained either with intracellular expression of full-length PA or with N-terminally truncated PA form, which lacks the signal peptide, is unable to provide protective immunity against B. anthracis STI anthrax infection (Duc et al. 2007).
Antibiotic-associated diarrhea induced by C. difficile has emerged as an important nosocomial infectious. Usually, the major virulence factors of these pathogens are the two toxins A and B (Jank and Aktories 2008). Given the importance of mucosal immunity in C. difficile infection, an illustrative example of how the CotB- and CotC-mediated spore display were used to obtain increased protection in a hamster against C. difficile infection when administered orally with the recombinant spores expressing the repeat domain of toxin A has been described previously (Permpoonpattana et al. 2011). Another recent report of Colenutt (2014) provided further evidence on the efficacy of spore display based-vaccine in inducing both systemic and mucosal responses. The delivery of this type of vaccine by a sublingual route resulted in a higher level of protection than that of an oral route. Interestingly, inactivated recombinant spores were more effective than live spores when delivered via the same route.
In addition to toxins A and B, another immunogenic fragment of FliD, the flagellar cap protein of C. difficile flagellin, used as a new vaccine candidate has recently been investigated (Negri et al. 2013). By fusion of the fragment of FliD to the C-terminus of CotB, CotC, CotG, or CotZ, successful display of FliD on the surface of B. subtilis spores can be achieved; however, in this reported example, the effectiveness of the recombinant spores surface-expressing CotB–GGGEAAAKGGG–FliD, CotC–FliD, CotG–FliD, or CotZ–FliD for developing an oral vaccine against C. difficile has yet to be examined in immunization of laboratory animals (Negri et al. 2013). Densitometric analysis shows that the amount of recombinant FliD protein in the spore coat was in the order of CotG–FliD (4.1 × 103 recombinant molecules per spore) > CotC–FliD (3.4 × 103) > CotZ–FliD (2.1 × 103) > CotB–FliD (6.9 × 102). A literature review by McKenney and Eichenberger (2012) suggests that the display efficiency of heterologous antigen or protein on the spore obtained with crust protein may theoretically be considerably higher than that achieved with the outer coat protein of B. subtilis spores. However, the results of the CotZ-mediated spore display for the expression of UreA and FliD did not completely support this hypothesis (Hinc et al. 2013; Negri et al. 2013). Apparently, the pattern of localization of fusion proteins within the spore coat may depend mainly on both the nature of target and anchor proteins as well as the characteristics of the fusion protein.
The selection of an adequate antigen could help in developing an efficient anti-H. pylori vaccine. Hinc et al. (2010b) attempted to create a vaccine by tethering UreA from H. acinonychis on the spores as a fusion to CotB, CotC, and CotG, respectively. Among the tested anchoring motifs, CotB showed the highest display efficiency of UreA. Although both CotC and CotG enabled UreA to be efficiently expressed, no fusion proteins were displayed on the spores. In addition to the failure of spore display of UreA, the CotG-based UreA fusion protein may suffer from partially processed. However, this observation seems to be in poor agreement in the literature on the use of CotG as an anchoring motif for the efficient surface expression of several different proteins, including streptavidin (Kim et al. 2005), β-galactosidase (Kwon et al. 2007), Neu5Ac aldolase (Xu et al. 2011a), and ω-transaminase (Hwang et al. 2011). Recently, another attempt has been made to improve the spore display-based delivery system in terms of immune response (Hinc et al. 2014). As reported, fused to the coat protein CotC, a fragment of subunit B from H. acinonychis urease (HaUreBF) was demonstrated to be presented on the surface of B. subtilis spores. Apart from the display of HaUreBF, the full-length ureB gene of H. acinonychis was expressed in vegetative cells. In addition, for the development of a more effective HaUreB vaccine, an appropriate adjuvant, human interleukin 2 (IL-2) displayed with CotB, was used. Results showed that mice immunized via oral route simultaneously with recombinant spores expressing HaUreBF and intracellular expressed HaUreB within vegetative cells developed cellular immune response at a higher level when co-administered with recombinant spores presenting IL-2 compared with mice vaccinated with a vaccine whose production was accomplished through conventional B. subtilis spore display (Hinc et al. 2014). Furthermore, the oral administration of recombinant spores presenting HaUreB together with IL-2-displaying spores or with aluminum hydroxide by healthy mice was shown to elicit a mixed Th1/Th17 polarized immune response. This response may provide added protection against this pathogen (Stasiłojć et al. 2015).
WSSV is currently the most serious viral pathogen in crustaceans globally. In a recent research, a highly immunogenic VP28, which is recognized as a promising vaccine candidate against WSSV (Akhila et al. 2014), was displayed on the surface of B. subtilis 168 spores by fusion to CotB or CotC (Ning et al. 2011). Oral administration of either recombinant spores displaying CotB–VP28 or CotC–VP28 in invertebrate crayfish generated a significant increase in survival rate, with levels of 37.9 and 44.8%, respectively, compared with 10.3% in crayfish orally immunized with wild-type spores (Ning et al. 2011). Similarly, the survival rate of Litopenaeus vannamei vaccinated with the recombinant spores surface-expressing CotB–VP28 was significantly higher than that of untreated animals after 14 days (Nguyen et al. 2014). Therefore, the administration of the recombinant spores displaying VP28 could strengthen the immune abilities of both crayfish and L. vannamei and increase their resistance to WSSV infection.
As previously noted, optimization of the promoter sequence is another approach to improve spore display efficiency. In a recent work done by Nguyen and Schumann (2014), two different β-D-thiogalactoside (IPTG)-inducible promoters P grac and P Sgrac were tested for the respective surface expression of α-amylase Q (AmyQ) from Bacillus amyloliquefaciens and GFPuv on the spore using CotB, CotC, and CotG as anchoring motifs. The P grac promoter was found to enable the display of both AmyQ and GFPuv on the spore surface. The expression level of CotB–AmyQ displayed on the spore coat under control of P Sgrac was the highest as compared to expression levels driven by P grac or P cotB , but their activities followed the order of P grac –CotB–AmyQ > P cotB –CotB–AmyQ > P Sgrac –CotB–AmyQ, postulating the existence of an enzymatically inactive form of AmyQ on the spore surface. Furthermore, the authors observed that only overproduced proteins among the three utilized coat proteins for displaying GFPuv tend to accumulate at different positions on the spore surface (Nguyen and Schumann 2014). The difference in arrangements may be a result of self-interaction within each overproduced coat protein and their tendency of localization on the spore coat.
More recently, by fusion of Bsβ–Gal to the C-terminus of each of CotB, CotC, CotG, CotX, CotY, or CotZ, an active enzyme could be displayed on the spore surface of B. subtilis 168 (Wang et al. 2015, 2016). The activity of spore-displayed Bsβ–Gal was in a sequence of CotG–Bsβ–Gal > CotX–Bsβ–Gal > CotB–Bsβ–Gal > CotZ–Bsβ–Gal > CotC–Bsβ–Gal > CotY–Bsβ–Gal. In addition, CotX–Bsβ–Gal was further tested for its transgalactosylation activity, yielding 8.8 g/L of lactulose from lactose and fructose. Hence, similar to CotG, CotX can exhibit considerable potential to target more bioactive molecules at the surface of B. subtilis spores. PhoA, an alkaline phosphatase of E. coli, is a disulfide bond-containing protein and becomes biologically active only when disulfide bonds can be formed. As shown by Richter et al. (2015), PhoA was correctly anchored on the B. subtilis spores as a fusion with either CotB or CotZ. The activity of spore-displayed PhoA was confirmed by enzyme assay. Further, analysis of the double-knockout experiment revealed that the deficient in both two proteins BdbC and BdbD exert no effect on the formation of disulfide bonds in recombinant spores surface-expressing PhoA. Collectively, these data prove that Bacillus spore display is a more suitable method for the immobilization of these disulfide-bonded-containing proteins.
Innovative B. subtilis spore display system
In all conventional B. subtilis spore display formats discussed so far, the system requires an anchor protein to target the displayable antigen or protein onto the spore surface. As stated above, this display format also presents certain limitations. However, researchers have been and have continue to invest considerable effort toward increasing B. subtilis spore display efficiency. With considerable increase in knowledge on spore structure and recent advances in biotechnology engineering, an innovative approach for direct display of native protein on B. subtilis spore without anchor protein as a scaffold was first proposed by Pan et al. (2014). In that report, the key element in determining the success of this native protein display system is the cry1Aa promoter, which is a σE- and σK-dependent promoter. When the gene of interest is placed under the sporulation-specific promoter, the target protein is preferably synthesized and displayed onto the spore surface. Through the use of this novel display strategy, two model enzymes, one being monomeric carboxymethylcellulase (CMCase) and the other being tetrameric β-galactosidase, can be successfully displayed on the spore surface in their respective native form, as verified by enzyme activity assays, flow cytometry, and immunogold electron microscopy experiments. Despite the demonstrated success in both CMCase and β-galactosidase, the general applicability of this promising display system along with the use of cry1Aa promoter for other proteins needs further investigation. As compared to non-recombinant spore display, the innovative B. subtilis spore display system offers clear advantages, namely, obviating the need for protein purification and preventing undesired detachment of displayed protein from the spore surface. This innovative B. subtilis spore display will not only widen the range of displayable proteins but also expand application fields.
Conclusions and future prospects
This review has highlighted several of the recent progress in understanding B. subtilis spore display. As a powerful tool, B. subtilis spore display conceivably offers broad possibilities in its future. However, current application of B. subtilis spore display is not yet feasible from laboratory-scale trial to commercialization. The very low display efficiency is a main barrier that blocks the development of B. subtilis spore display. Further studies on the exploration of a wider variety of anchor protein present a promising approach to address this great challenge. In addition, the display performance of heterologous antigen or protein can be potentially increased by approaches such as redesigning the surface structure of spore coat, optimization of coat protein folding and assembly, directed evolution of coat protein, increasing the copy number of the gene cassettes, and construction of a versatile vector. Meanwhile, the excellent availability of genetic manipulation tool for B. subtilis will help in the redesign and exploitation of metabolic pathways for improved target antigen or protein expression in display host cells. Importantly, several issues involved in innovative B. subtilis spore display system should also be addressed. For instance, how does this native protein display work, what about display efficiency when compared with the conventional display format, and can the display serve as drug delivery vehicle and withstand in harsh conditions?
However, application aspects of B. subtilis spore display will require additional directions. One possibility is to catalyze multi-step transformation for directly producing bio-based chemicals by applying B. subtilis spores that co-display a combination of the appropriate enzymes. An exciting field is in the targeted drug delivery system for therapy in diverse diseases. Another approach is devoted to the continuing development of more effective vaccine with unique properties. In view of the past innovation in the field, B. subtilis spore display tool is expected to be commercialized for industrial applications.
References
Akhila DS, Mani MK, Rai P, Condon K, Owens L, Karunasagar I (2014) Antisense RNA mediated protection from white spot syndrome virus (WSSV) infection in Pacific white shrimp Litopenaeus vannamei. Aquaculture 435:306–309
Amuguni H, Tzipori S (2012) Bacillus subtilis: a temperature resistant and needle free delivery system of immunogens. Hum Vaccin Immunother 8:979–986
Batista MT, Souza RD, Paccez JD, Luiz WB, Ferreira EL, Cavalcante RC, Ferreira RC, Ferreira LC (2014) Gut adhesive Bacillus subtilis spores as a platform for the mucosal delivery of antigens. Infect Immun 82:1414–1423
Cao Y, Li Z, Yue Y, Song N, Peng L, Wang L, Lu X (2013) Construction and evaluation of a novel Bacillus subtilis spores based enterovirus 71 vaccine. J Appl Biomed 11:105–113
Chen H, Tian R, Ni Z, Zhang Q, Zhang T, Chen Z, Chen K, Yang S (2015a) Surface display of the thermophilic lipase Tm1350 on the spore of Bacillus subtilis by the CotB anchor protein. Extremophiles 19:799–808
Chen H, Zhang T, Jia J, Ake V, Tian R, Ni Z, Chen Z, Chen K, Yang S (2015b) Expression and display of a novel thermostable esterase from Clostridium thermocellum on the surface of Bacillus subtilis using the CotB anchor protein. J Ind Microbiol Biotechnol 42:1439–1448
Chen H, Zhang T, Sun T, Ni Z, Le Y, Tian R, Chen Z, Zhang C (2015c) Clostridium thermocellum nitrilase expression and surface display on Bacillus subtilis spores. J Mol Microbiol Biotechnol 25:381–387
Chen H, Chen Z, Ni Z, Tian R, Zhang T, Jia J, Chen K, Yang S (2016) Display of Thermotoga maritima MSB8 nitrilase on the spore surface of Bacillus subtilis using out coat protein CotG as the fusion partner. J Mol Catal B Enzym 123:73–80
Ciabattini A, Parigi R, Isticato R, Oggioni MR, Pozzi G (2004) Oral priming of mice by recombinant spores of Bacillus subtilis. Vaccine 22:4139–4143
Colenutt C (2014) Use of Bacillus subtilis spores in treatments for Clostridium difficile infection. Dissertation, University of London
Crameri A, Whitehorn EA, Tate E, Stemmer WP (1996) Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol 14:315–319
Cutting SM (2011) Bacillus probiotics. Food Microbiol 28:214–220
Cutting SM, Vander-Horn PB (1990) Genetic analysis. In: Harwood CR, Cutting SM (eds) Molecular biological methods for Bacillus. Wiley, Chichester, pp 27–60
Cutting SM, Hong HA, Baccigalupi L, Ricca E (2009) Oral vaccine delivery by recombinant spore probiotics. Int Rev Immunol 28:487–505
D’Apice L, Sartorius R, Caivano A, Mascolo D, Del Pozzo G, Di Mase DS, Ricca E, Li Pira G, Manca F, Malanga D, De Palma R, De Berardinis P (2007) Comparative analysis of new innovative vaccine formulations based on the use of procaryotic display systems. Vaccine 25:1993–2000
Driks A (1999) Bacillus subtilis spore coat. Microbiol Mol Biol Rev 63:1–20
Duc LH, Cutting SM (2003a) Bacterial spores as heat stable vaccine vehicles. Expert Opin Biol Ther 3:1263–1270
Duc LH, Hong HA, Fairweather N, Ricca E, Cutting SM (2003b) Bacterial spores as vaccine vehicles. Infect Immun 71:2810–2818
Duc LH, Hong HA, Atkins HS, Flick-Smith HC, Durrani Z, Rijipkema S, Titball RW, Cutting SM (2007) Immunization against anthrax using Bacillus subtilis spores expressing the anthrax protective antigen. Vaccine 25:346–355
Feng F, Hu P, Chen L, Tang Q, Lian C, Yao Q, Chen K (2013) Display of human proinsulin on the Bacillus subtilis spore surface for oral administration. Curr Microbiol 67:1–8
Gao C, Xu X, Zhang X, Che B, Ma C, Qiu J, Tao F, Xu P (2011) Chemoenzymatic synthesis of N-acetyl-D-neuraminic acid from N-acetyl-D-glucosamine by using the spore surface-displayed N-acetyl-D-neuraminic acid aldolase. Appl Environ Microbiol 77:7080–7083
Henriques AO, Moran CP Jr (2007) Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol 61:555–588
Hinc K, Ghandili S, Karbalaee G, Shali A, Noghabi K, Ricca E, Ahmadian G (2010a) Efficient binding of nickel ions to recombinant Bacillus subtilis spores. Res Microbiol 161:757–764
Hinc K, Isticato R, Dembek M, Karczewska J, Iwanicki A, Peszyńska-Sularz G, De Felice M, Obuchowski M, Ricca E (2010b) Expression and display of UreA of Helicobacter acinonychis on the surface of Bacillus subtilis spores. Microb Cell Factories 9:2
Hinc K, Iwanicki A, Obuchowski M (2013) New stable anchor protein and peptide linker suitable for successful spore surface display in B. subtilis. Microb Cell Factories 12:22
Hinc K, Stasiłojć M, Piątek I, Peszyńska-Sularz G, Isticato R, Ricca E, Obuchowski M, Iwanicki A (2014) Mucosal adjuvant activity of IL-2 presenting spores of Bacillus subtilis in a murine model of Helicobacter pylori vaccination. PLoS One 9:e95187
Hoang TH, Hong HA, Clark GC, Titball RW, Cutting SM (2008) Recombinant Bacillus subtilis expressing the Clostridium perfringens alpha toxoid is a candidate orally delivered vaccine against necrotic enteritis. Infect Immun 76:5257–5265
Hong HA, Duc LH, Cutting SM (2006) The use of bacterial spore formers as probiotics. FEMS Microbiol Rev 29:813–835
Hong HA, Huang JM, Khaneja R, Hiep LV, Urdaci MC, Cutting SM (2008) The safety of Bacillus subtilis and Bacillus indicus as food probiotics. J Appl Microbiol 105:510–520
Hosseini-Abari A, Kim BG, Lee SH, Emtiazi G, Kim W, Kim JH (2016) Surface display of bacterial tyrosinase on spores of Bacillus subtilis using CotE as an anchor protein. J Basic Microbiol 56:1–7
Huang JM, La Ragione RM, Cooley WA, Todryk S, Cutting SM (2008) Cytoplasmic delivery of antigens, by Bacillus subtilis enhances Th1 responses. Vaccine 26:6043–6052
Huang JM, Hong HA, Van Tong H, Hoang TH, Brisson A, Cutting SM (2010) Mucosal delivery of antigens using adsorption to bacterial spores. Vaccine 28:1021–1030
Hwang BY, Kim BG, Kim JH (2011) Bacterial surface display of a co-factor containing enzyme, ω-transaminase from Vibrio fluvialis using the Bacillus subtilis spore display system. Biosci Biotechnol Biochem 75:1862–1865
Hwang BY, Pan JG, Kim BG, Kim JH (2013) Functional display of active tetrameric β-galactosidase using Bacillus subtilis spore display system. J Nanosci Nanotechnol 13:2313–2319
Imamura D, Kuwana R, Takamatsu H, Watabe K (2011) Proteins involved in formation of the outermost layer of Bacillus subtilis spores. J Bacteriol 193:4075–4080
Isticato R, Ricca E (2014) Spore surface display. Microbiol Spect 2:TBS-0011-2012
Isticato R, Cangiano G, Tran HT, Ciabattini A, Medaglini D, Oggioni MR, De Felice M, Del Pozzo G, Ricca E (2001) Surface display of recombinant proteins on Bacillus subtilis spores. J Bacteriol 183:6294–6301
Isticato R, Di Mase DS, Mauriello EM, De Felice M, Ricca E (2007) Amino terminal fusion of heterologous proteins to CotC increases display efficiencies in the Bacillus subtilis spore system. BioTechniques 42:151–156
Iwanicki A, Piątek I, Stasiłojć M, Grela A, Łęga T, Obuchowski M, Hinc K (2014) A system of vectors for Bacillus subtilis spore surface display. Microb Cell Factories 13:30
Jank T, Aktories K (2008) Structure and mode of action of clostridial glucosylating toxins: the ABCD model. Trends Microbiol 16:222–229
Kim J, Schumann W (2009) Display of proteins on Bacillus subtilis endospores. Cell Mol Life Sci 66:3127–3136
Kim JH, Kim BG, Choi SK, Jung HC, Pan JG (2004) Method for expression of proteins on spore surface. US Patent 7, 582,426 B2
Kim JH, Lee CS, Kim BG (2005) Spore-displayed streptavidin: a live diagnostic tool in biotechnology. Biochem Biophy Res Commun 331:210–214
Kim JH, Roh C, Lee CW, Kyung D, Choi SK, Jung HC, Pan JG, Kim BG (2007) Bacterial surface display of GFPuv on Bacillus subtilis spores. J Microbiol Biotechnol 17:677–680
Knecht LD, Pasini P, Daunert S (2011) Bacterial spores as platforms for bioanalytical and biomedical applications. Anal Bioanal Chem 400:977–989
Kunst F, Ogasawara N, Moszer I et al (1997) The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256
Kwon SJ, Jung HC, Pan JG (2007) Transgalactosylation in a water-solvent biphasic reaction system with β-galactosidase displayed on the surfaces of Bacillus subtilis spores. Appl Environ Microbiol 73:2251–2256
Lee SY, Choi JH, Xu Z (2003) Microbial cell-surface display. Trends Biotechnol 21:45–52
Li L, Hu X, Wu Z, Xiong S, Zhou Z, Wang X, Lu F, Yu X (2009) Immunogenicity of self-adjuvanticity oral vaccine candidate based on use of Bacillus subtilis spore displaying Schistosoma japonicum 26 KDa GST protein. Parasitol Res 105:1643–1651
Li Q, Ning D, Wu C (2010) Surface display of GFP using CotX as a molecular vector on Bacillus subtilis spores. Chinese J Biotechnol 26:264–269
Li G, Tang Q, Chen H, Yao Q, Ning D, Chen K (2011) Display of Bombyx mori nucleopolyhedrovirus GP64 on the Bacillus subtilis spore coat. Curr Microbiol 62:1368–1373
Lian C, Zhou Y, Feng F, Chen L, Tang Q, Yao Q, Chen K (2014) Surface display of human growth hormone on Bacillus subtilis spores for oral administration. Curr Microbiol 68:463–471
Liu Y, Li S, Xu H, Wu L, Xu Z, Liu J, Feng X (2014) Efficient production of D-tagatose using a food-grade surface display system. J Agric Food Chem 62:6756–6762
Mao L, Jiang S, Li G, He Y, Chen L, Yao Q, Chen K (2012) Surface display of human serum albumin on Bacillus subtilis spores for oral administration. Curr Microbiol 64:545–551
Mauriello EM, Duc LH, Isticato R, Cangiano G, Hong HA, De Felice M, Ricca E, Cutting SM (2004) Display of heterologous antigens on the Bacillus subtilis spore coat using CotC as a fusion partner. Vaccine 22:1177–1187
Mauriello EM, Cangiano G, Maurano F, Saggese V, De Felice M, Rossi M, Ricca E (2007) Germination-independent induction of cellular immune response by Bacillus subtilis spores displaying the C fragment of the tetanus toxin. Vaccine 25:788–793
McKenney PT, Eichenberger P (2012) Dynamics of spore coat morphogenesis in Bacillus subtilis. Mol Microbiol 83:245–260
McKenney PT, Driks A, Eichenberger P (2013) The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nature 11:33–44
Medaglini D, Ciabattini A, Spinosa MR, Maggi T, Marcotte H, Oggioni MR, Pozzi G (2001) Immunization with recombinant Streptococcus gordonii expressing tetanus toxin fragment C confers protection from lethal challenge in mice. Vaccine 19:1931–1939
Mou C, Zhu L, Xing X, Lin J, Yang Q (2016) Immune responses induced by recombinant Bacillus subtilis expressing the spike protein of transmissible gastroenteritis virus in pigs. Antivir Res 131:74–84
Negri A, Potocki W, Iwanicki A, Obuchowski M, Hinc K (2013) Expression and display of Clostridium difficile protein FliD on the surface of Bacillus subtilis spores. J Med Microbiol 62:1379–1385
Nguyen QA, Schumann W (2014) Use of IPTG-inducible promoters for anchoring recombinant proteins on the Bacillus subtilis spore surface. Protein Expr Purif 95:67–76
Nguyen VAT, Huynh HA, Hoang TV, Ninh NT, Pham ATH, Nguyen HA, Phan TN, Cutting SM (2013) Killed Bacillus subtilis spores expressing streptavidin: a novel carrier of drugs to target cancer cells. J Drug Target 21:528–541
Nguyen ATV, Pham CK, Pham HTT, Pham HL, Nguyen AH, Dang LT, Huynh HA, Cutting SM, Phan TN (2014) Bacillus subtilis spores expressing the VP28 antigen: a potential oral treatment to protect Litopenaeus vannamei against white spot syndrome. FEMS Microbiol Lett 358:202–208
Ning D, Leng X, Li Q, Xu W (2011) Surface-displayed VP28 on Bacillus subtilis spores induce protection against white spot syndrome virus in crayfish by oral administration. J Appl Microbiol 111:1327–1336
Oggioni MR, Ciabattini A, Cuppone AM, Pozzi G (2003) Bacillus spores for vaccine delivery. Vaccine 21:96–101
Ozin AJ, Samford CS, Henriques AO, Moran CP Jr (2001) SpoVID guides SafA to the spore coat in Bacillus subtilis. J Bacteriol 183:3041–3049
Pan JG, Kim EJ, Yun CH (2012) Bacillus spore display. Trends Biotechnol 30:610–612
Pan JG, Choi SK, Jung HC, Kim EJ (2014) Display of native proteins on Bacillus subtilis spores. FEMS Microbiol Lett 358:209–217
Permpoonpattana P, Hong HA, Phetcharaburanin J, Huang JM, Cook J, Fairweather NF, Cutting SM (2011) Immunization with Bacillus spores expressing toxin A peptide repeats protects against infection with Clostridium difficile strains producing toxins A and B. Infect Immun 79:2295–2302
Piggot PJ, Hilbert DW (2004) Sporulation of Bacillus subtilis. Curr Opin Microbiol 7:579–586
Potot S, Serra CR, Henriques AO, Schyns G (2010) Display of recombinant proteins on Bacillus subtilis spores, using a coat-associated enzyme as the carrier. Appl Environ Microbiol 76:5926–5933
Qu H, Xu Y, Sun H, Lin J, Yu J, Tang Z, Shen J, Liang C, Li S, Chen W, Li X, Wu Z, Huang Y, Yu X (2014a) Systemic and local mucosal immune responses induced by orally delivered Bacillus subtilis spore expressing leucine aminopeptidase 2 of Clonorchis sinensis. Parasitol Res 113:3095–3103
Qu Y, Wang J, Zhang Z, Shi S, Li D, Shen W, Shen E, Zhou J (2014b) Catalytic transformation of HODAs using an efficient meta-cleavage product hydrolase-spore surface display system. J Mol Catal B Enzym 102:204–210
Richter A, Kim W, Kim JH, Schumann W (2015) Disulfide bonds of proteins displayed on spores of Bacillus subtilis can occur spontaneously. Curr Microbiol 71:156–161
Schallmey M, Singh A, Ward OP (2004) Developments in the use of Bacillus species for industrial production. Can J Microbiol 50:1–17
Setlow P (2006) Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol 101:514–525
Sibley L, Reljic R, Radford DS, Huang JM, Hong HA, Cranenburgh RM, Cutting SM (2014) Recombinant Bacillus subtilis spores expressing MPT64 evaluated as a vaccine against tuberculosis in the murine model. FEMS Microbiol Lett 358:170–179
Smith GP (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315–1317
Stasiłojć M, Hinc K, Peszyńska-Sularz G, Obuchowski M, Iwanicki A (2015) Recombinant Bacillus subtilis spores elicit Th1/Th17-polarized immune response in a murine model of Helicobacter pylori vaccination. Mol Biotechnol 57:685–691
Tavassoli S, Hinc K, Iwanicki A, Obuchowski M, Ahmadian G (2013) Investigation of spore coat display of Bacillus subtilis β-galactosidase for developing of whole cell biocatalyst. Arch Microbiol 195:197–202
Unnikrishnan M, Rappuoli R, Serruto D (2012) Recombinant bacterial vaccines. Curr Opin Immun 24:337–342
Uyen NQ, Hong HA, Cutting SM (2007) Enhanced immunisation and expression strategies using bacterial spores as heat-stable vaccine delivery vehicles. Vaccine 25:356–365
Valdez A, Yepiz-Plascencia G, Ricca E, Olmos J (2014) First Litopenaeus vannamei WSSV 100% oral vaccination protection using CotC::Vp26 fusion protein displayed on Bacillus subtilis spores surface. J Appl Microbiol 117:347–357
Wang N, Chang C, Yao Q, Li G, Qin L, Chen L, Chen K (2011) Display of Bombyx mori alcohol dehydrogenases on the Bacillus subtilis spore surface to enhance enzymatic activity under adverse conditions. PLoS One 6:e21454
Wang X, Chen W, Tian Y, Mao Q, Lv X, Shang M, Li X, Yu X, Huang Y (2014) Surface display of Clonorchis sinensis enolase on Bacillus subtilis spores potentializes an oral vaccine candidate. Vaccine 32:1338–1345
Wang H, Yang R, Hua X, Zhao W, Zhang W (2015) Functional display of active β-galactosidase on Bacillus subtilis spores using crust proteins as carriers. Food Sci Biotechnol 24:1755–1759
Wang H, Yang R, Hua X, Zhang W, Zhao W (2016) An approach for lactulose production using the CotX-mediated spore-displayed β-galactosidase as a biocatalyst. J Microbiol Biotechnol 26:1267–1277
Wu CH, Mulchandani A, Chen W (2008) Versatile microbial surface-display for environmental remediation and biofuels production. Trends Microbiol 16:181–188
Xu X, Gao C, Zhang X, Che B, Ma C, Qiu J, Tao F, Xu P (2011a) Production of N-acetyl-D-neuraminic acid by use of an efficient spore surface display system. Appl Environ Microbiol 77:3197–3201
Xu Z, Qing Y, Li S, Feng X, Xu H, Ouyang P (2011b) A novel L-arabinose isomerase from Lactobacillus fermentum CGMCC2921 for D-tagatose production: gene cloning, purification and characterization. J Mol Catal B Enzym 70:1–7
Yu J, Chen T, Xie Z, Liang P, Qu H, Shang M, Mao Q, Ning D, Tang Z, Shi M, Zhou L, Huang Y, Yu X (2015) Oral delivery of Bacillus subtilis spore expressing enolase of Clonorchis sinensis in rat model: induce systemic and local mucosal immune responses and has no side effect on liver function. Parasitol Res 114:2499–2505
Yuan Y, Feng F, Chen L, Yao Q, Chen K (2014) Surface display of Acetobacter pasteurianus AdhA on Bacillus subtilis spores to enhance ethanol tolerance for liquor industrial potential. Eur Food Res Technol 238:285–293
Zhao G, Miao Y, Guo Y, Qiu H, Sun S, Kou Z, Yu H, Li J, Chen Y, Jiang S, Du L, Zhou Y (2014) Development of a heat-stable and orally delivered recombinant M2e-expressing B. subtilis spore-based influenza vaccine. Hum Vaccin Immunother 10:3649–3658
Zhou Z, Xia H, Hu X, Huang Y, Ma C, Chen X, Hu F, Xu J, Lu F, Wu Z, Yu X (2008a) Immunogenicity of recombinant Bacillus subtilis spores expressing Clonorchis sinensis tegumental protein. Parasitol Res 102:293–297
Zhou Z, Xia H, Hu X, Huang Y, Li Y, Li L, Ma C, Chen X, Hu F, Xu J, Lu F, Wu Z, Yu X (2008b) Oral administration of a Bacillus subtilis spore-based vaccine expressing Clonorchis sinensis tegumental protein 22.3 kDa confers protection against Clonorchis sinensis. Vaccine 26:1817–1825
Zhou Z, Gong S, Li XM, Yang Y, Guan R, Zhou S, Yao S, Xie Y, Ou Z, Zhao J, Liu Z (2015) Expression of Helicobacter pylori urease B on the surface of Bacillus subtilis spores. J Med Microbiol 64:104–110
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported by the project from the Scientific Research & Development Fund of Jiyang College, Zhejiang Agriculture and Forestry University (JY2014RC009, JYMS1409) and Food Science and Engineering the most important discipline of Zhejiang province (JYTsp20142101).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Wang, H., Wang, Y. & Yang, R. Recent progress in Bacillus subtilis spore-surface display: concept, progress, and future. Appl Microbiol Biotechnol 101, 933–949 (2017). https://doi.org/10.1007/s00253-016-8080-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-8080-9