Abstract
The targeting and anchoring of heterologous proteins and peptides to the outer surface of bacteriophages and cells is becoming increasingly important, and has been employed as a tool for fundamental and applied research in microbiology, molecular biology, vaccinology, and biotechnology. Less known are endospores or spores produced by some Gram-positive species. Spores of Bacillus subtilis are surrounded by a spore coat on their outside, and a few proteins have been identified being located on the outside layer and have been successfully used to immobilize antigens and some other proteins and enzymes. The major advantage of spores over the other published systems is their synthesis within the cytoplasm of the bacterial cell. Therefore, any heterologous protein to be anchored on the outside does not have to cross any membrane. Furthermore, spores are extremely resistant against high temperature, irradiation and many chemicals, and can be stored for many years at room temperature.
Similar content being viewed by others
Introduction
Surface display is a powerful technique that uses natural microbial functional components to express heterologous peptides and proteins on the exterior of phages or cells. The first surface expression system was developed by George P. Smith who used the gene III of the filamentous phage M13 [1] for display. This gene codes for the protein pIII which is present in 5–8 copies near one end of the phage particle [2, 3]. The pIII protein consists of two domains separated by a linker region. Smith published the pioneering idea to insert parts of the coding region of an endonuclease gene in-frame in the linker region followed by the identification of the recombinant phages using appropriate antibodies in a technique termed biopanning [1]. This technique is now well known as surface display of peptides and proteins on the surface not only of phages but also on whole cells of bacteria and yeast. Besides the filamentous phages, display systems have also been developed for bacteriophages, λ, e.g., [4–6], T4, e.g., [6–8], and T7 [9]. The concept of using naturally occurring surface proteins as an anchor for targeting proteins of interest (often called passenger proteins), with a distinct function at the surface of phages and cells summarized as bioparticles, offers a broad range of applications in many different areas of bioscience, including the screening of novel binding partners, delivery of vaccines and drugs, production of active enzymes and antibodies for cleanup of industrial and environmental pollution, and their use as biosensors, biocatalysts for bioconversion, and screening of peptides libraries [6–8]. All these systems share a common theme, targeting recombinant proteins to the phage or cell surface by constructing gene fusions using sequences from membrane-anchored domains of surface or phage coat proteins.
The polypeptides displayed at the particle surface are freely accessible to the substrate or binding partner in activity and binding studies. Furthermore, proteins have been proven to be more stable when connected to a bioparticle rather than as free, diffusible molecules, and this makes it unnecessary to purify these proteins. There are several excellent review articles dealing with the application of surface display using bacterial cells and bacteriophages [10–20].
When filamentous phages or bacterial cells are used for surface display, the chimeric proteins have to cross at least one membrane, the cytoplasmic membrane. For obvious reasons, not every chimeric protein is able to cross this membrane. Those proteins which either fold fast in the cytoplasm or which contain hydrophobic patches provoking entrapment in the membrane are refractory to translocation through the cytoplasmic membrane. All these problems can be prevented by using endospores formed by Bacillus and related aerobic endospore-forming bacteria (a group of some 200 species, distributed over 25 genera) as well as by Clostridium as a strategy to survive during unfavorable conditions. Since these spores are formed within the sporulating cells, the mother cells, Bacillus offers four advantages. First, the chimeric proteins do not have to cross the cytoplasmic membrane and also fast folding passenger proteins should be anchored on the spore surface. Furthermore, the full set of ATP-dependent molecular chaperones is present to assist the correct folding of the passenger protein if necessary. Third, proteins being multimers to become active or proteins that need an intracellular co-factor (for example, transaminase with pyrodoxal phosphate as a co-factor) are other challenging targets for spore surface display. And fourth, it should even be possible to obtain formation of disulfide bonds by providing the appropriate conditions within the cytoplasm as has been published for E. coli [21, 22]. In the following chapters, we will first review the most relevant data leading to spore formation in B. subtilis, the spore composition and the germination process. Then, the current state of spore surface display will be reviewed.
Formation of the B. subtilis endospore
When confronted by nutrient depletion, the normally rod-shaped B. subtilis cells produce an oval dormant cell, the spore. The molecular and biochemical reactions underlying the genetic program resulting in spore formation have been studied most extensively in B. subtilis, but all members of the two groups Bacillus and Clostridium produce endospores according to a program which is similar to that of B. subtilis [23–25]. Bacterial endospores are commonly found in any soil and water environment, but they can colonize virtually any habitat. They are entirely distinct from the vegetative cell, possessing several molecular and cellular structures seen nowhere else in nature. These unique components contribute to the spore’s most striking characteristics. It is metabolically inactive, highly resilient to environmental assault, and stable for extreme periods of time. Spores are the most resistant form of life on earth, and they can withstand extremes of heat, radiation, chemical assault, and time [26, 27]. Viable spores were recovered from the gut of a bee fossilized in Dominican amber for an estimated 25–40 million years [28]. But the spore can sense the reappearance of even minute amounts of nutrients in the environment, and respond by converting back to a vegetative growing cell [29]. Very recently, it has been published that muropeptides also act as potent germinants [30]. This process leading to the conversion of spores to vegetative cells is called germination (see below).
Five different kinases, KinA through KinE [31], sense so far unknown starvation factors, autophosphorylate, and transfer the phosphoryl group through two proteins, Spo0F and Spo0B, to the master regulator Spo0A [32]. This process is called phosphorelay, and Spo0A~P acts both as a transcriptional repressor and activator to regulate transcription of a total of 121 genes [33]. A major function of Spo0A~P is to activate expression of genes encoding two sporulation-specific sigma factors. As sporulation proceeds, there is an unequal cell division that generates a large mother cell and a smaller forespore (Fig. 1). The plasma membrane of the mother cell then grows around the forespore, generating an engulfed forespore surrounded by two membranes. Activation and/or synthesis of additional sigma factors in a compartment-specific manner then drive different patterns of gene expression in the mother cell and in the forespore compartment. The subsequent synthesis of a thick peptidoglycan cortex between the outer and inner forespore membrane is accompanied by a large decrease in the volume and water content of the forespore protoplast, and there is also a decrease in the forespore pH [34]. The forespore later takes up tremendous amounts of pyridine-2,6-dicarboxylic acid (dipicolinic acid, DPA) that has been synthesized in the mother cell, which decreases the forespore water content even further (see [35–37] for recent reviews).
Stages of sporulation. a Vegetative cell in the phase 0; b cell at stage II where the asymmetric septum has been formed; c cell at stage III where the cytoplasmic membrane of the mother cell has engulfed the forespore; d cell at stage IV where formation of the coat has already started; the spore is about to be released from the lysed mother cell. Figure adapted from [104]
Composition of the B. subtilis endospore
The Bacillus spore consists of three concentric compartments separated by two membranes (Fig. 2). The central part has been designated the spore core. It contains one copy of the bacterial chromosome and plasmid copies if present in the vegetative cell. The DNA molecules are complexed with small acid-soluble proteins (SASPs), the predominant proteins of the core making up as much as 20% of the total spore protein [38]. The most abundant SASPs fall into one of three classes, based on sequence comparisons and biochemical properties: α, β, and γ [39]. The SASPs protect spores from a wide range of damaging agents, including heat, UV radiation, nucleases, hydroxyl radicals, formaldehyde, nitrous acid nucleases and desiccation [40]. The α/β-type SASPs protect the spore by binding to and saturating its DNA [41]. These proteins do not possess any known DNA-binding motifs, and their interaction with DNA is relatively non-specific. Upon binding to DNA, the α/β-type SASPs alter DNA conformation locally, stiffening the DNA and changing it from a B to an A conformation [42]. Besides the SASPs, the dehydrated state of the core and ions such as DPA, mainly as calcium chelate [43], also play critical roles. A second function of the SASPs is to serve as a source of amino acids after proteolysis, toward the end of germination (see below). Furthermore, the normal cellular cytoplasmic components, from RNA, enzymes to ribosomes, are present, but all in an inactive state.
Structure of the spore showing the different compartments. The innermost compartment, the core, contains the DNA and is filled with small acid-soluble proteins that saturate the DNA and protect it and all the other components present in the cytoplasm in a largely dehydrated state. The spore core is surrounded by two membranes sandwiching a specialized peptidoglycan called cortex keeping the water activity relatively low. The spore coat consists of two layers and is composed of about 70 different proteins. Pathogenic and some saprophytic Bacilli possess an additional layer called exosporium consisting of a paracrystalline basal layer and a hair-like nap. A thin section electron micrograph showing a spore with its exosporium has been presented [108]
The inner membrane of the dormant spore is in an unusual environment and has been described as ‘semi-crystalline’, only regaining the fluidity characteristics of a normal cell membrane when the spore germinates [44]. It has a low permeability to small molecules, perhaps even water [45]. The low permeability of this membrane is one factor in the resistance of spores to some chemicals, in particular DNA-damaging chemicals [40].
The surface of the inner spore membrane is the site of assembly of a thin layer of peptidoglycan called the primordial germ cell wall, similar in composition to the vegetative cell wall that serves as the primordial wall of the newly formed vegetative cell following spore germination (Fig. 2). The outer prespore membrane is the site of assembly of a second, thicker, and chemically distinct layer of peptidoglycan called the spore cortex, which is essential for the attainment and maintenance of the dehydrated state of the spore core, for spore mineralization and for dormancy [27, 46, 47]. The cortex beneath the outer spore membrane is composed predominantly of peptidoglycan with a structure similar to that of peptidoglycan in growing cells. However, it contains two modifications: muramic acid-δ-lactam and muramic acid linked only to alanine.
The third compartment, the spore coat, is a complex, multilayered assemblage of more than 50 proteins that encases the spore and helps to protect it from noxious environmental agents [46]. It is composed out of two major layers [26, 27]: a lightly staining, finally striated inner coat composed of several fine lamellae and a more darkly staining coarsely layered outer coat, made up of several thicker sublayers. The more than 50 different coat proteins range in size from about 6 to about 70 kDa, some of which have been subject to detailed analysis [46, 48, 49]. The assembling coat is tethered to this membrane by the 26-amino-acid SpoVM peptide, which is believed to form an amphipathic helix. It has been shown that proper localization of SpoVM is dependent on SpoIVA, a morphogenetic protein that forms the basement layer of the spore coat [50]. Conversely, proper localization of SpoIVA is dependent on SpoVM, and this mutual dependence is mediated in part by an amino acid side-chain located near the extreme C-terminus of SpoIVA and an amino acid side-chain on the hydrophilic face of the SpoVM helix. SpoIVA both binds and hydrolyses ATP and self-assembles into cable-like structures depending on ATP hydrolysis [51]. The coat apparently has a variety of functions. It has long been known to serve as a barrier against entry of large toxic molecules, such as a lysozyme, and to have a role in germination [26]. The most recent identification of a germination protein [52] in the coat should help to clarify its role in germination, and the discovery of coat protein oxidases [53–55] points to additional active roles that have yet to be elucidated.
Most coat proteins appear to be dispensable for the coat’s barrier functions, because the deletion of any one of the corresponding genes has no detectable consequence. Two important exceptions are SpoIVA and CotE. SpoIVA resides between the coat and the cortex [56, 57]. As this location suggests, SpoIVA connects the coat to the spore surface; without it, the coat is built but not attached to the spore [47, 58, 59]. CotE is positioned at the inner coat/outer coat interface [56]. It is responsible for the assembly of the entire outer coat as well as several inner coat proteins [60–63].
Germination of the B. subtilis endospore
Spores germinate in response to an environmental signal, and the most common germinants are amino acids, sugars or ribosides. This germination process uses components already established in the spore during its formation, and is essentially a biophysical and degradative one [64]. The germinant has to first penetrate the coat and cortex layers of the spore before coming in contact with the germinant receptors [65] located in the inner spore membrane [66]. In B. subtilis, these receptors are encoded by three homologous tricistronic operons termed gerA, gerB and gerK which are expressed in the forespore late in sporulation [64, 67]. While the GerA receptor recognizes l-alanine, the GerB and GerK receptors are involved for germination in the presence of a mixture of asparagine, glucose, fructose and K+ (AGFK). When spores become committed to germination, the following reactions will take place [64, 67].
First, the downstream consequences of germination-receptor interaction include a rapid efflux of monovalent cations (H+, Na+ and K+) and Zn2+, where the release of H+ elevates the core pH from ~6.5 to 7.7. Second, the large depot of DPA (about 10% of the spore dry weight) and its associated divalent cations (mainly Ca2+) will be released and replaced by water. Third, lytic enzymes that hydrolyze the specialized peptidoglycan of the cortex are activated, leading to full rehydration [52, 68–70].
In B. subtilis, one of the two germination-specific lytic enzymes (CwlJ) is localized at the coat–cortex interface, and can be activated artificially by high concentrations of external Ca2+-DPA, which might also be the natural activating signal for this enzyme [71]. The other major lytic enzyme, SleB, is present in the spore inner membrane and in the outer layers [68]. Both enzymes, CwlJ and SleB, play redundant roles in the degradation of the spore’s peptidoglycan cortex during germination. Therefore, the germination frequency in a double knockout is reduced by five orders of magnitude [72]. A similar reduction can be obtained by using the gerD-cwlB double knockout mutant. The SASPs are degraded by a specific protease, releasing DNA from its complexed state [67]. The metabolizing and energized germinated spore can then resume RNA, protein, and DNA synthesis in the outgrowth phase [73]. This further increase in core hydration will result in protein mobility and activation of enzyme activities. All these events take place without detectable energy metabolism. The return of enzymatic activities in the spore core allows initiation of spore metabolism, followed by macromolecular synthesis finally converting the germinated spore into a growing cell, and this period has been designated spore outgrowth.
Anchoring of passenger proteins on the endospore surface of B. subtilis
The spore-display system is based on the construction of gene fusions between heterologous DNA and a B. subtilis gene coding for a component of the spore coat, where the spore coat protein has been shown to be located on its outside. Three different spore coat proteins have been used to far, CotB, CotC, and CotG. All three proteins are tyrosine-rich coat components present as multimers within the spore coat [74, 75], and they are mainly if not exclusively associated with the outer coat [75–78]. The cotB gene encodes a 46-kDa polypeptide (CotB-46) which is posttranslationally converted into a form of about 66 kDa (CotB-66) [76]. CotC forms multimeric species with itself and the nearly identical CotU protein at the surface [75, 79]. CotG has a central region formed by nine tandem repeats of a lysine-, serine-, and arginine-rich motif [78]. While in the case of CotC and CotG the full-length versions are used as an anchoring motif, with CotB a shorter version is used [77].
Until now, major applications using B. subtilis spores are focused on delivery of surface-expressed antigens using CotB and CotC as anchoring motifs. Initial developments in this field have been summarized in three reviews [80–82]. Since then, additional antigens, detailed results about vaccination, immunogenicity, and the fate of spores within the animals have been published. The CotG protein has been used to anchor different enzymes on the spore surface. In the following sections, we will focus on various spore-based display systems and their applications as well as the initial developments in this field.
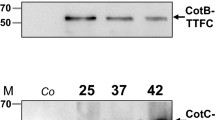
The first report of spore surface display was done with CotB as an anchoring motif and the coding region of TTFC (C-terminal fragment of the tetanus toxin), a well-characterized and highly immunogenic model antigen [83]. The 459-amino-acid TTFC was fused to the N-terminus, the truncated C-terminus of CotB, and was inserted into the middle of CotB. All three fusion proteins exhibited a similar surface location as confirmed by flow cytometry [84]. Next, spores displaying the C-terminal CotB-TTFC fusion were used for oral and intranasal immunization of mice. It turned out that these recombinant spores generated both a mucosal (IgA) and a systemic (IgG) response in the murine model [85]. In a further report, mice were inoculated intragastrically with spores displaying the CotB-TTFC, and a significant high serum TTFC-specific IgG response was stimulated [86]. The responses after delivery of the TTFC antigen expressed either on the spore surface or in the germinating spore were compared [87]. Antigen on the spore surface produced a more pronounced, but less rapid, response with a type 1 T cell response bias.
The protective antigen (PA) from a toxin-producing B. anthracis strain is a key protein for the interaction of a lethal factor and the host membrane, and neutralization of PA and prevention of its binding to the host cell surface was a main target for vaccine development. PA has been shown to induce protection against infection by that strain [88]. Besides oral and mucosal delivery, parenteral delivery of B. subtilis spores carrying PA was tested [89]. Spores carrying different (31, 48, and 91 kDa) domains of the PA at the C-terminal end of CotB or CotC were delivered into six mice through the intra-peritoneal route, and were very effective to confer protective immunity using an in vitro toxin neutralization assay and a challenge experiment with the latter showing protection to a lethal dose of B. anthracis spore.
Besides CotB, the CotC protein has also been used as an anchoring motif for the display of TTFC and the LTB (heat-labile toxin of E. coli) antigen [90]. In both cases, the antigen-displaying spores provoked an immune response in mice, when they were delivered by intra-peritoneal injection or by oral administration. A Chinese group published two related reports for the development of spore-based vaccines against human clonorchiasis using CotC as an anchoring motif [91, 92]. Caused by infection with the nematode of C. sinensis, clonorchiasis is endemic in southern China [93], Korea, and other southeast Asian countries. By large-scale sequencing of a C. sinensis cDNA library, they had identified three full-length cDNAs encoding three putative tegumental proteins (TP20.8, TP22.3, TP31.8), which could be the most susceptible target for vaccines because of their importance for the host response and parasite survival. Among them, they performed parallel study with TP20.8 [94] and TP22.3 [91]. In both cases, when antigen-displaying spores were delivered orally, they reported an elevated secretory level of IgA, which aggregated pathogens, inhibited their motility, and prevented their adherence to epithelial cells. However, the protection rate against metacercariae challenge had no significant differences compared with those of control groups, when spore-displaying TP20.8 was delivered. Only spore-expressing TP22.3 could induce a significant level of protection evidenced as reduced worm burden in the vaccinated rats (44.7%), suggesting that TP20.8 is a real membrane protein with a primary transmembrane helix as an interesting vaccine candidate.
In a recent publication [95], comparison of antigenicity and immunogenicity of a T helper epitope pep23 expressed in three different vaccine delivery systems was carried out. In an experiment performed by using a specific T cell hybridoma and by priming mononuclear cells isolated from the venous blood of human donors, spore display system with fusion at the N-terminal of CotC was less effective when compared to filamentous bacteriophage fd display system or cell surface display system based on E2 protein from the PDH complex of the B. stearothermophilus system. However, considering the safety and ease of mass production, Bacillus spores have many valuable applications in mucosal administration.
In a completely different application using CotG and CotE as anchoring motives, β-galactosidase of E. coli, the most utilized reporter protein in bioscience and biotechnology, was surface-expressed on B. subtilis spores [96]. β-Galactosidase is a very large protein (116 kDa per monomer), and active only as a tetramer and known to be toxic to the host cell when translocation was tried using the Sec pathway of host cells causing membrane jamming. β-Galactosidase was fused to the C-terminal ends of CotG and CotE each. Its surface expression was verified by flow cytometry and by a protease accessibility test. This result demonstrated the advantage of a spore-based display system compared to the vegetative cell-based approach in that it has obviously no limit in size (116 × 4 = 464 kDa) and in the multimeric nature of the target protein.
Besides hydrolyzing lactose to glucose and galactose, β-galactosidase can also catalyze a transglycosylation reaction in water-organic solvent biphasic reaction systems [97]. With the spore surface-anchored β-galactosidase, alkyl-β-galactosides, a group of nonionic surfactants, which exhibit antimicrobial activity and are used as drug intermediates, could be produced [98]. Solvent stability of surface expressed β-galactosidase, especially in ethyl ether, toluene, ethyl acetate, and acetonitrile was significantly increased as compared to the free form of soluble β-galactosidase. And the thermostability of surface-expressed β-galactosidase was also increased compared to free soluble β-galactosidase. Through this approach, the problem of low stability of the enzyme in solvents and a strong limitation in mass transfer between water and solvent phases separating enzymes and substrates could be eliminated.
In another application, streptavidin was immobilized on the spore surface [77]. Streptavidin is a 60-kDa protein produced by Streptomyces avidinii and binds biotin very tightly with a K d of 10−15. Active streptavidin is composed of four identical molecules with a molecular weight of 15 kDa each, and four active sites for biotin binding exist in the active tetramer. Biotin can be easily conjugated to various biomolecules, such as proteins, nucleic acids, and carbohydrates, and spore-displayed streptavidin can be widely used for the detection of biotin-conjugated biomolecules. But the high G + C content of the DNA sequence (69%) and sequestration of intracellular biotin, which is lethal to the host bacteria, and tetramer formation of streptavidin makes it difficult to express streptavidin intracellularly or to secrete it in its active form. Using the CotG anchoring motif, they generated streptavidin-coated B. subtilis spores, and its localization was confirmed by anti-streptavidin antibodies. Furthermore, the biological activity of surface-expressed streptavidin was directly confirmed by Biotin-FITC conjugate by flow cytometry, and streptavidin-coated spores can directly recognize biotin-FITC on glass surfaces, confirmed by fluorescence microscopy. This application could provide a simple and superior platform compared to the B. thuringiensis micropatterns (see below) [99], eliminating antibody and biotin-protein A treatment on biotin-patterned surfaces.
In a third application, GFPuv, a variant of the green fluorescent protein, was surface-displayed on B. subtilis spores using the CotG anchoring motif [100]. Two different GFPuv display vectors (with and without the coding region of cotG) were constructed to examine the precise role of CotG as an anchoring motif for the display of GFPuv. As to be expected, only spores expressing the fusion protein exhibited fluorescence due to surface-anchored GFPuv. This application offers the interesting possibility to use GFPuv for the identification of additional spore coat proteins present on the outside such as CotU to be used as anchoring motif.
Anchoring of proteins on the spore surface of other endospore-formers
Many Bacillus species can be found in the soil, where the majority of these are saprophytes. But three species, B. cereus, B. anthracis, and B. thuringiensis, are pathogens of animals and insects. The spore architecture of these pathogens, as well as of some of the saprophytes, differs from that of B. subtilis in possessing an additional outermost layer called exosporium [101] (Fig. 2). The exosporium surrounds the spore coat and consists of a paracrystalline basal layer and an external hair-like nap. In B. anthracis, the basal layer contains more than a dozen different integral proteins, and the filaments of the hair-like nap are formed mainly by a single collagen-like protein called BclA [102] and the less frequent protein BclB [103]. Sequences near the N-terminus of both glycoproteins are responsible for incorporation of BclA and BclB into the exosporium layer of the spore. They were used to immobilize GFP onto the spore surface [104], suggesting a new vaccine delivery system based on inactivated spores of B. anthracis.
Spores of B. thuringiensis contain the 130-kDa protoxin from the Cry1Ac subgroup as a major component of the spore coat layer, which forms originally parasporal crystal inclusion outside the exosporium [105]. It has been proposed that the N-terminal part of the protoxin is exposed on the spore surface, while the C-terminal region anchors the protein inside the spore coat [106]. Two different passenger proteins have been used to replace the N-terminal end, and the chimeric proteins were successfully anchored on the spore surface: GFP and a single-chain antibody (scFv) recognizing a chemical compound [107]. The advantages of B. thuringiensis spores for surface display are the higher sporulation rate (98–100 vs 60–70% for B. subtilis) and the finding that the protoxin is not essential, while in B. subtilis both the native Cot proteins and the Cot fusion proteins are anchored on the surface of the spores.
Micropatterns are spatially well-defined, two-dimensional microstructures of, e.g., cells on a solid surface such as a glass slide, which can serve as sensors/detectors. Here, micropatterns of B. thuringiensis spores have been prepared [99]. First, EGFP was fused to the InhA protein, and these spores were then treated with anti-GFP antibodies, followed by addition of protein A-biotin. These complexes were added to a glass slide with immobilized streptavidin. The spores were analyzed under the microscope and exhibited an ellipsoidal shape. In addition, the authors could show that the immobilized spores were still able to germinate when the glass slides were placed with the upside-down configuration on LB agar plates [99]. All the major applications of proteins displayed on spore surface are listed in Table 1.
Concluding remarks
Spore display is an interesting alternative to phage and cell display. It offers four important advantages when compared to the other systems: (1) the fusion proteins do not have to cross a membrane; (2) ATP-dependent molecular chaperones are present and can assist correct folding of the fusion proteins; (3) spore display allows oligomerization of proteins as shown in the case of β-galactosidase; and (4) there is no obvious codon bias. Spores can be prepared at a high rate (~1010/ml) from flask cultures and can be stored for indefinite times after purification at room temperature. This allows the establishment of combinatorial libraries of, e.g., single chain libraries. One open question is why not all spores express the passenger protein on their surface. When spores are analyzed by flow cytometry, sometimes only about 50% of the spores exhibit a signal ([92], and our unpublished results). Using GFP as passenger protein, additional spore coat proteins can be identified being surface exposed such as CotU [79].
References
Smith GP (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315–1317
Pratt D, Tzagoloff H, Beaudoin J (1969) Conditional lethal mutants of the small filamentous coliphage M13. II: two genes for coat proteins. Virology 39:42–53
Grant RA, Lin TC, Konigsberg W, Webster RE (1981) Structure of the filamentous bacteriophage fl: location of the A, C, and D minor coat proteins. J Biol Chem 256:539–546
Maruyama IN, Maruyama HI, Brenner S (1994) λfoo: a λ phage vector for the expression of foreign proteins. Proc Natl Acad Sci USA 91:8273–8277
Dunn IS (1996) Total modification of the bacteriophage lambda tail tube major subunit protein with foreign peptides. Gene 183:15–21
Hoess RH (2002) Bacteriophage lambda as a vehicle for peptide and protein display. Curr Pharm Biotechnol 3:23–28
Efimov VP, Nepluev IV, Mesyanzhinov VV (1995) Bacteriophage T4 as a surface display vector. Virus Genes 10:173–177
Ren Z, Black LW (1998) Phage T4 SOC and HOC display of biologically active, full-length proteins on the viral capsid. Gene 215:439–444
Houshmand H, Froman G, Magnusson G (1999) Use of bacteriophage T7 displayed peptides for determination of monoclonal antibody specificity and biosensor analysis of the binding reaction. Anal Biochem 268:363–370
Lee SY, Choi JH, Xu Z (2003) Microbial cell-surface display. Trends Biotechnol 21:45–52
Wernerus H, Stahl S (2004) Biotechnological applications for surface-engineered bacteria. Biotechnol Appl Biochem 40:209–228
Georgiou G, Stathopoulos C, Daugherty PS, Nayak AR, Iverson BL, Curtiss RIII (1997) Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat Biotechnol 15:29–34
Cossart P, Jonquieres R (2000) Sortase, a universal target for therapeutic agents against gram-positive bacteria? Proc Natl Acad Sci USA 97:5013–5015
Benhar I (2001) Biotechnological applications of phage and cell display. Biotechnol Adv 19:1–33
Samuelson P, Gunneriusson E, Nygren PA, Stahl S (2002) Display of proteins on bacteria. J Biotechnol 96:129–154
Jose J, Meyer TF (2007) The autodisplay story, from discovery to biotechnical and biomedical applications. Microbiol Mol Biol Rev 71:600–619
Saleem M, Brim H, Hussain S, Arshad M, Leigh MB, Zia H (2008) Perspectives on microbial cell surface display in bioremediation. Biotechnol Adv 26:151–161
Daugherty PS (2007) Protein engineering with bacterial display. Curr Opin Struct Biol 17:474–480
Sidhu SS, Koide S (2007) Phage display for engineering and analyzing protein interaction interfaces. Curr Opin Struct Biol 17:481–487
Wu CH, Mulchandani A, Chen W (2008) Versatile microbial surface-display for environmental remediation and biofuels production. Trends Microbiol 16:181–188
Bessette PH, Åslund F, Beckwith J, Georgiou G (1999) Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci USA 96:13703–13708
DeLisa MP, Tullman D, Georgiou G (2003) Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc Natl Acad Sci USA 100:6115–6120
Holt SC, Leadbetter ER (1969) Comparative ultrastructure of selected aerobic spore-forming bacteria: a freeze-etching study. Bacteriol Rev 33:346–378
Priest FG (1993) Systematics and ecology of Bacillus. In: Sonenshein AL, Hoch JA, Losick R (eds) Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC, pp 3–16
Sonenshein AL (2000) Endospore-forming bacteria: an overview. In: Brun YV, Shimkets LJ (eds) Prokaryotic development. American Society for Microbiology, Washington, DC, pp 133–150
Aronson AI, Fitz-James PC (1976) Structure and morphogenesis of the bacterial spore coat. Bacteriol Rev 40:360–402
Driks A (1999) Bacillus subtilis spore coat. Microbiol Mol Biol Rev 63:1–20
Cano RJ, Borucki MK (1995) Revival and identification of bacterial spores in 25- to 40-million-year-old Dominican amber. Science 268:1060–1064
Atrih A, Foster SJ (1999) The role of peptidoglycan structure and structural dynamics during endospore dormancy and germination. Antonie Van Leeuwenhoek 75:299–307
Shah IM, Laaberki MH, Popham DL, Dworkin J (2008) A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135:486–496
Jiang M, Shao W, Perego M, Hoch JA (2000) Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol 38:535–542
Hoch JA (2000) Two-component and phosphorelay signal transduction. Curr Opin Microbiol 3:165–170
Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, Losick R (2003) The Spo0A regulon of Bacillus subtilis. Mol Microbiol 50:1683–1701
Popham DL (2002) Specialized peptidoglycan of the bacterial endospore: the inner wall of the lockbox. Cell Mol Life Sci 59:426–433
Sonenshein AL (2000) Control of sporulation initiation in Bacillus subtilis. Curr Opin Microbiol 3:561–566
Piggot PJ, Hilbert DW (2004) Sporulation of Bacillus subtilis. Curr Opin Microbiol 7:579–586
Errington J (2003) Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1:117–126
Setlow P, Waites WM (1976) Identification of several unique, low-molecular-weight basic proteins in dormant spores of clastridium bifermentans and their degradation during spore germination. J Bacteriol 127:1015–1017
Setlow P (1988) Small, acid-soluble spore proteins of Bacillus species: structure, synthesis, genetics, function, and degradation. Annu Rev Microbiol 42:319–338
Setlow P (2006) Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol 101:514–525
Setlow P (1995) Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu Rev Microbiol 49:29–54
Mohr SC, Sokolov NVHA, He C, Setlow P (1991) Binding of small acid-soluble spore proteins from Bacillus subtilis changes the conformation of DNA from B to A. Proc Natl Acad Sci USA 88:77–81
Paidhungat M, Setlow P (2000) Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J Bacteriol 182:2513–2519
Stewart GS, Eaton MW, Johnstone K, Barrett MD, Ellar DJ (1980) An investigation of membrane fluidity changes during sporulation and germination of Bacillus megaterium K.M. measured by electron spin and nuclear magnetic resonance spectroscopy. Biochim Biophys Acta 600:270–290
Cortezzo DE, Setlow P (2005) Analysis of factors that influence the sensitivity of spores of Bacillus subtilis to DNA damaging chemicals. J Appl Microbiol 98:606–617
Henriques AO, Moran CP Jr (2000) Structure and assembly of the bacterial endospore coat. Methods 20:95–110
Piggot PJ, Coote JG (1976) Genetic aspects of bacterial endospore formation. Bacteriol Rev 40:908–962
Driks A (2002) Proteins of the spore coat. In: Sonenshein AL, Hoch JA, Losick R (eds) Bacillus subtilis and its closest relatives. Am Soc Microbiol, Washington, DC, pp 527–536
Kim H, Hahn M, Grabowski P, McPherson DC, Otte MM, Wang R, Ferguson CC, Eichenberger P, Driks A (2006) The Bacillus subtilis spore coat protein interaction network. Mol Microbiol 59:487–502
Ramamurthi KS, Clapham KR, Losick R (2006) Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol 62:1547–1557
Ramamurthi KS, Losick R (2008) ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol Cell 31:406–414
Bagyan I, Setlow P (2002) Localization of the cortex lytic enzyme CwlJ in spores of Bacillus subtilis. J Bacteriol 184:1219–1224
Lai EM, Phadke ND, Kachman MT, Giorno R, Vazquez S, Vazquez JA, Maddock JR, Driks A (2003) Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J Bacteriol 185:1443–1454
Martins LO, Soares CM, Pereira MM, Teixeira M, Costa T, Jones GH, Henriques AO (2002) Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J Biol Chem 277:18849–18859
Hullo MF, Moszer I, Danchin A, Martin-Verstraete I (2001) CotA of Bacillus subtilis is a copper-dependent laccase. J Bacteriol 183:5426–5430
Driks A, Roels S, Beall B, Moran CP, Losick R Jr (1994) Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev 8:234–244
Pogliano K, Harry E, Losick R (1995) Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol Microbiol 18:459–470
Roels S, Driks A, Losick R (1992) Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol 174:575–585
Stevens CM, Daniel R, Illing N, Errington J (1992) Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J Bacteriol 174:586–594
Little S, Driks A (2001) Functional analysis of the Bacillus subtilis morphogenetic spore coat protein CotE. Mol Microbiol 42:1107–1120
Bauer T, Little S, Stover AG, Driks A (1999) Functional regions of the Bacillus subtilis spore coat morphogenetic protein CotE. J Bacteriol 181:7043–7051
Takamatsu H, Chikahiro Y, Kodama T, Koide H, Kozuka S, Tochikubo K, Watabe K (1998) A spore coat protein, CotS, of Bacillus subtilis is synthesized under the regulation of sigmaK and GerE during development and is located in the inner coat layer of spores. J Bacteriol 180:2968–2974
Zilhao R, Naclerio G, Henriques AO, Baccigalupi L, Moran CP, Ricca E Jr (1999) Assembly requirements and role of CotH during spore coat formation in Bacillus subtilis. J Bacteriol 181:2631–2633
Moir A, Corfe BM, Behravan J (2002) Spore germination. Cell Mol Life Sci 59:403–409
Paidhungat M, Setlow P (1999) Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant d-alanine. J Bacteriol 181:3341–3350
Hudson KD, Corfe BM, Kemp EH, Feavers IM, Coote PJ, Moir A (2001) Localization of GerAA and GerAC germination proteins in the Bacillus subtilis spore. J Bacteriol 183:4317–4322
Paidhungat M, Setlow P (2002) Spore germination and outgrowth. In: Sonenshein AL, Hoch JA, Losick R (eds) Bacillus subtilis and its closest relatives. American Society for Microbiology, Washington, DC, pp 537–548
Chirakkal H, O’Rourke M, Atrih A, Foster SJ, Moir A (2002) Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology 148:2383–2392
Boland FM, Atrih A, Chirakkal H, Foster SJ, Moir A (2000) Complete spore-cortex hydrolysis during germination of Bacillus subtilis 168 requires SleB and YpeB. Microbiology 146:57–64
Shimamoto S, Moriyama R, Sugimoto K, Miyata S, Makino S (2001) Partial characterization of an enzyme fraction with protease activity which converts the spore peptidoglycan hydrolase (SleC) precursor to an active enzyme during germination of Clostridium perfringens S40 spores and analysis of a gene cluster involved in the activity. J Bacteriol 183:3742–3751
Paidhungat M, Ragkousi K, Setlow P (2001) Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca(2+)-dipicolinate. J Bacteriol 183:4886–4893
Setlow B, Melly E, Setlow P (2001) Properties of spores of Bacillus subtilis blocked at an intermediate stage in spore germination. J Bacteriol 183:4894–4899
Horsburgh MJ, Thackray PD, Moir A (2001) Transcriptional responses during outgrowth of Bacillus subtilis endospores. Microbiology 147:2933–2941
Zilhao R, Serrano M, Isticato R, Ricca E, Moran CP, Henriques AO Jr (2004) Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J Bacteriol 186:1110–1119
Isticato R, Esposito G, Zilhao R, Nolasco S, Cangiano G, De FM, Henriques AO, Ricca E (2004) Assembly of multiple CotC forms into the Bacillus subtilis spore coat. J Bacteriol 186:1129–1135
Donovan W, Zheng LB, Sandman K, Losick R (1987) Genes encoding spore coat polypeptides from Bacillus subtilis. J Mol Biol 196:1–10
Kim JH, Lee CS, Kim BG (2005) Spore-displayed streptavidin: a live diagnostic tool in biotechnology. Biochem Biophys Res Commun 331:210–214
Sacco M, Ricca E, Losick R, Cutting S (1995) An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J Bacteriol 177:372–377
Isticato R, Pelosi A, Zilhao R, Baccigalupi L, Henriques AO, De FM, Ricca E (2008) CotC–CotU heterodimerization during assembly of the Bacillus subtilis spore coat. J Bacteriol 190:1267–1275
Ricca E, Cutting SM (2003) Emerging applications of bacterial spores in nanobiotechnology. J Nanobiotechnol 1:6–16
Oggioni MR, Ciabattini A, Cuppone AM, Pozzi G (2003) Bacillus spores for vaccine delivery. Vaccine 21(Suppl 2):96–101
Ferreira LC, Ferreira RC, Schumann W (2005) Bacillus subtilis as a tool for vaccine development: from antigen factories to delivery vectors. An Acad Bras Cienc 77:113–124
Medaglini D, Ciabattini A, Spinosa MR, Maggi T, Marcotte H, Oggioni MR, Pozzi G (2001) Immunization with recombinant Streptococcus gordinii expressing tetanus toxin fragment C confers protection from lethal challenge in mice. Vaccine 19:1931–1939
Isticato R, Cangiano G, Tran HT, Ciabattini A, Medaglini D, Oggioni MR, De FM, Pozzi G, Ricca E (2001) Surface display of recombinant proteins on Bacillus subtilis spores. J Bacteriol 183:6294–6301
Duc IH, Hong HA, Fairweather N, Ricca E, Cutting SM (2003) Bacterial spores as vaccine vehicles. Infect Immun 71:2810–2818
Ciabattini A, Parigi R, Isticato R, Oggioni MR, Pozzi G (2004) Oral priming of mice by recombinant spores of Bacillus subtilis. Vaccine 22:4139–4143
Uyen NQ, Hong HA, Cutting SM (2007) Enhanced immunisation and expression strategies using bacterial spores as heat-stable vaccine delivery vehicles. Vaccine 25:356–365
Flick-Smith HC, Eyles JE, Hebdon R, Waters EL, Beedham RJ, Stagg TJ, Miller J, Alpar HO, Baillie LW, Williamson ED (2002) Mucosal or parenteral administration of microsphere-associated Bacillus anthracis protective antigen protects against anthrax infection in mice. Infect Immun 70:2022–2028
Duc IH, Hong HA, Atkins HS, Flick-Smith HC, Durrani Z, Rijpkema S, Titball RW, Cutting SM (2007) Immunization against anthrax using Bacillus subtilis spores expressing the anthrax protective antigen. Vaccine 25:346–355
Mauriello EM, Duc IH, Isticato R, Cangiano G, Hong HA, De FM, Ricca E, Cutting SM (2004) Display of heterologous antigens on the Bacillus subtilis spore coat using CotC as a fusion partner. Vaccine 22:1177–1187
Zhou Z, Xia H, Hu X, Huang Y, Li Y, Li L, Ma C, Chen X, Hu F, Xu J (2008) Oral administration of a Bacillus subtilis spore-based vaccine expressing Clonorchis sinensis tegumental protein 22.3 kDa confers protection against Clonorchis sinensis. Vaccine 26:1817–1825
Zhou Z, Xia H, Hu X, Huang Y, Ma C, Chen X, Hu F, Xu J, Lu F, Wu Z (2008) Immunogenicity of recombinant Bacillus subtilis spores expressing Clonorchis sinensis tegumental protein. Parasitol Res 102:293–297
Zhang RL, Gao ST, Huang DN, Zhang SX, Cheng JQ, Fu YC (2007) Epidemiological study on Clonorchis sinensis infection in Shenzhen area of Zhujiang Delta in China. Parasitol Res 101:179–183
Zhou Z, Hu X, Huang Y, Hu H, Ma C, Chen X, Hu F, Xu J, Lu F, Wu Z (2007) Molecular cloning and identification of a novel Clonorchis sinensis gene encoding a tegumental protein. Parasitol Res 101:737–742
D’Apice L, Sartorius R, Caivano A, Mascolo D, Del PG, Di Mase DS, Ricca E, Li PG, Manca F, Malanga D (2007) Comparative analysis of new innovative vaccine formulations based on the use of procaryotic display systems. Vaccine 25:1993–2000
Ban JG., Choi SG., Jung HC., Kim BG, Kim JH (2003) Method for expression of proteins on spore surface. KR20030065534. South-Korea. Patent
Mori T, Fujita S, Okahata Y (1997) Transglycosylation in a two-phase aqueous-organic system with catalysis by a lipid-coated beta-d-galactosidase. Carbohydr Res 298:65–73
Kwon SJ, Jung HC, Pan JG (2007) Transgalactosylation in a water-solvent biphasic reaction system with beta-galactosidase displayed on the surfaces of Bacillus subtilis spores. Appl Environ Microbiol 73:2251–2256
Park TJ, Lee KB, Lee SJ, Park JP, Lee ZW, Choi SK, Jung HC, Pan JG, Lee SY, Choi IS (2004) Micropatterns of spores displaying heterologous proteins. J Am Chem Soc 126:10512–10513
Kim JH, Roh C, Lee CW, Kyung D, Choi SK, Jung HC, Pan JG, Kim BG (2007) Bacterial surface display of GFP(uv) on Bacillus subtilis spores. J Microbiol Biotechnol 17:677–680
Henriques O, Moran CP Jr (2007) Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol 61:555–588
Sylvestre P, Couture-Tosi E, Mock M (2002) A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol Microbiol 45:169–178
Thompson BM, Waller LN, Fox KF, Fox A, Stewart GC (2007) The BclB glycoprotein of Bacillus anthracis is involved in exosporium integrity. J Bacteriol 189:6704–6713
Thompson BM, Stewart GC (2008) Targeting of the BclA and BclB proteins to the Bacillus anthracis spore surface. Mol Microbiol 70:421–434
Short JA, Walker PD, Thomson RO, Somerville HJ (1974) The fine structure of Bacillus finitimus and Bacillus thuringiensis spores with special reference to the location of crystal antigen. J Gen Microbiol 84:261–276
Du C, Nickerson KW (1996) Bacillus thuringiensis HD-73 spores have surface-localized Cry1Ac Toxin: physiological and pathogenic consequences. Appl Environ Microbiol 62:3722–3726
Du C, Chan WC, McKeithan TW, Nickerson KW (2005) Surface display of recombinant proteins on Bacillus thuringiensis spores. Appl Environ Microbiol 71:3337–3341
Driks A (2002) Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol 10:251–254
Acknowledgments
This work was supported by a research fund from KICOS to Junehyung Kim and from PT-DLR to Wolfgang Schumann.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, J., Schumann, W. Display of proteins on Bacillus subtilis endospores. Cell. Mol. Life Sci. 66, 3127–3136 (2009). https://doi.org/10.1007/s00018-009-0067-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-009-0067-6