Abstract
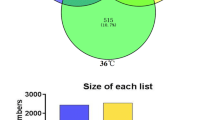
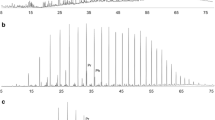
Bacterial communities in both aqueous and oil phases of water-flooded petroleum reservoirs were characterized by molecular analysis of bacterial 16S rRNA genes obtained from Shengli Oil Field using DNA pyrosequencing and gene clone library approaches. Metagenomic DNA was extracted from the aqueous and oil phases and subjected to polymerase chain reaction amplification with primers targeting the bacterial 16S rRNA genes. The analysis by these two methods showed that there was a large difference in bacterial diversity between the aqueous and oil phases of the reservoir fluids, especially in the reservoirs with lower water cut. At a high phylogenetic level, the predominant bacteria detected by these two approaches were identical. However, pyrosequencing allowed the detection of more rare bacterial species than the clone library method. Statistical analysis showed that the diversity of the bacterial community of the aqueous phase was lower than that of the oil phase. Phylogenetic analysis indicated that the vast majority of sequences detected in the water phase were from members of the genus Arcobacter within the Epsilonproteobacteria, which is capable of degrading the intermediates of hydrocarbon degradation such as acetate. The oil phase of reservoir fluid samples was dominated by members of the genus Pseudomonas within the Gammaproteobacteria and the genus Sphingomonas within the Alphaproteobacteria, which have the ability to degrade crude oil through adherence to hydrocarbons under aerobic conditions. In addition, many anaerobes that could degrade the component of crude oil were also found in the oil phase of reservoir fluids, mainly in the reservoir with lower water cut. These were represented by Desulfovibrio spp., Thermodesulfovibrio spp., Thermodesulforhabdus spp., Thermotoga spp., and Thermoanaerobacterium spp. This research suggested that simultaneous analysis of DNA extracted from both aqueous and oil phases can facilitate a better understanding of the bacterial communities in water-flooded petroleum reservoirs.






Similar content being viewed by others
References
Abbasnezhad H, Foght JM, Gray MR (2011) Adhesion to the hydrocarbon phase increases phenanthrene degradation by Pseudomonas fluorescens LP6a. Biodegradation 22(3):485–496. doi:10.1007/s10532-010-9421-5
Aeckersberg F, Bak F, Widdel F (1991) Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch Microbiol 156(1):5–14. doi:10.1007/BF00418180
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402. doi:10.1093/nar/25.17.3389
Angiuoli SV, Matalka M, Gussman A, Galens K, Vangala M, Riley DR, Arze C, White JR, White O, Fricke WF (2011) CloVR: a virtual machine for automated and portable sequence analysis from the desktop using cloud computing. BMC Bioinforma 12:356. doi:10.1186/1471-2105-12-356
Ashelford KE, ChuzhanovaNA FJC, Jones AJ, Weightman AJ (2005) At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71(12):7724–7736. doi:10.1128/AEM.71.12.7724-7736.2005
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2006) New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72(9):5734–5741. doi:10.1128/aem.00556-06
Beeder J, Torsvik T, Lien T (1995) Thermodesulforhabdus norvegicus gen. nov., sp. nov., a novel thermophilic sulfate-reducing bacterium from oil field water. Arch Microbiol 164(5):331–336. doi:10.1007/bf02529979
Brown L (2013) The use of microorganisms to enhance oil recovery. In: James S (ed) Enhanced oil recovery field case studies. Gulf Professional Publishing, Boston, pp 561–580
Callbeck CM, Dong X, Chatterjee I, Agrawal A, Caffrey SM, Sensen CW, Voordouw G (2011) Microbial community succession in a bioreactor modeling a souring low-temperature oil reservoir subjected to nitrate injection. Appl Microbiol Biotechnol 91(3):799–810. doi:10.1007/s00253-011-3287-2
Cephas KD, Kim J, Mathai RA, Barry KA, Dowd SE, Meline BS, Swanson KS (2011) Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing. PLoS ONE 6(8):e23503. doi:10.1371/journal.pone.0023503
Chen H, Boutros P (2011) VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinforma 12(1):35. doi:10.1186/1471-2105-12-35
Davidova IA, Suflita JM (2005) Enrichment and isolation of anaerobic hydrocarbon-degrading bacteria. Methods Enzymol 397:17–34. doi:10.1016/s0076-6879(05)97002-x
Ehrenreich P, Behrends A, Harder J, Widdel F (2000) Anaerobic oxidation of alkanes by newly isolated denitrifying bacteria. Arch Microbiol 173(1):58–64. doi:10.1007/s002030050008
Fedorovich V, Knighton MC, Pagaling E, Ward FB, Free A, Goryanin I (2009) Novel electrochemically active bacterium phylogenetically related to Arcobacter butzleri, isolated from a microbial fuel cell. Appl Environ Microbiol 75(23):7326–7334. doi:10.1128/aem.01345-09
Fry JC, Horsfield B, Sykes R, Cragg BA, Heywood C, Kim GT, Mangelsdorf K, Mildenhall DC, Rinna J, Vieth A, Zink K-G, Sass H, Weightman AJ, Parkes RJ (2009) Prokaryotic populations and activities in an interbedded coal deposit, including a previously deeply buried section (1.6–2.3 km) above ∼ 150 ma basement rock. Geomicrobiol J 26(3):163–178. doi:10.1080/01490450902724832
Gieg LM, Duncan KE, Suflita JM (2008) Bioenergy production via microbial conversion of residual oil to natural gas. Appl Environ Microbiol 74(10):3022–3029. doi:10.1128/aem.00119-08
Grabowski A, Nercessian O, Fayolle F, Blanchet D, Jeanthon C (2005) Microbial diversity in production waters of a low-temperature biodegraded oil reservoir. FEMS Microbiol Ecol 54(3):427–443. doi:10.1016/j.resmic.2005.03.009
Gray ND, Sherry A, Grant RJ, Rowan AK, Hubert CRJ, Callbeck CM, Aitken CM, Jones DM, Adams JJ, Larter SR, Head IM (2011) The quantitative significance of Syntrophaceae and syntrophic partnerships in methanogenic degradation of crude oil alkanes. Environ Microbiol 13(11):2957–2975. doi:10.1111/j.1462-2920.2011.02570.x
Guan J, Xia L-P, Wang L-Y, Liu J-F, Gu J-D, Mu B-Z (2013) Diversity and distribution of sulfate-reducing bacteria in four petroleum reservoirs detected by using 16S rRNA and dsrAB genes. Int Biodeterior Biodegrad 76:58–66. doi:10.1016/j.ibiod.2012.06.021
Hubert CRJ, Oldenburg TBP, Fustic M, Gray ND, Larter SR, Penn K, Rowan AK, Seshadri R, Sherry A, Swainsbury R, Voordouw G, Voordouw JK, Head IM (2012) Massive dominance of Epsilonproteobacteria in formation waters from a Canadian oil sands reservoir containing severely biodegraded oil. Environ Microbiol 14(2):387–404. doi:10.1111/j.1462-2920.2011.02521.x
Jones DM, Head IM, Gray ND, Adams JJ, Rowan AK, Aitken CM, Bennett B, Huang H, Brown A, Bowler BFJ, Oldenburg T, Erdmann M, Larter SR (2008) Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451(7175):176–180. doi:10.1038/nature06484
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N (2009) A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 3(4):442–453. doi:10.1038/ismej.2008.127
Kertesz M, Kawasaki A (2010) Hydrocarbon-degrading Sphingomonads: Sphingomonas, Sphingobium, Novosphingobium, and Sphingopyxis. In: Timmis KN (ed) Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, pp 1693–1075
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kobayashi H, Endo K, Sakata S, Mayumi D, Kawaguchi H, Ikarashi M, Miyagawa Y, Maeda H, Sato K (2012) Phylogenetic diversity of microbial communities associated with the crude-oil, large-insoluble-particle and formation-water components of the reservoir fluid from a non-flooded high-temperature petroleum reservoir. J Biosci Bioeng 113(2):204–210. doi:10.1016/j.jbiosc.2011.09. 015
Korenblum E, Souza DB, Penna M, Seldin L (2012) Molecular analysis of the bacterial communities in crude oil Samples from two Brazilian offshore petroleum platforms. Int J Microbiol 2012:156537. doi:10.1155/2012/156537
Kröber M, Bekel T, Diaz NN, Goesmann A, Jaenicke S, Krause L, Miller D, Runte KJ, Viehöver P, Pühler A, Schlüter A (2009) Phylogenetic characterization of a biogas plant microbial community integrating clone library 16S-rDNA sequences and metagenome sequence data obtained by 454-pyrosequencing. J Biotechnol 142(1):38–49. doi:10.1016/j.jbiotec.2009.02.010
Kryachko Y, Dong X, Sensen C, Voordouw G (2012) Compositions of microbial communities associated with oil and water in a mesothermic oil field. Anton Leeuw 101(3):493–506. doi:10.1007/s10482-011-9658-y
Li H, Yang S-Z, Mu B-Z, Rong Z-F, Zhang J (2006) Molecular analysis of the bacterial community in a continental high-temperature and water-flooded petroleum reservoir. FEMS Microbiol Lett 257(1):92–98. doi:10.1111/j.1574-6968.2006.00149.x
Li H, Yang S-Z, Mu B-Z (2007a) Phylogenetic diversity of the Archaeal community in a continental high-temperature, water-flooded petroleum reservoir. Curr Microbiol 55(5):382–388. doi:10.1007/s00284-007-9002-y
Li H, Yang S-Z, Mu B-Z, Rong Z-F, Zhang J (2007b) Molecular phylogenetic diversity of the microbial community associated with a high-temperature petroleum reservoir at an offshore oilfield. FEMS Microbiol Ecol 60(1):74–84. doi:10.1111/j.1574-6941.2006.00266.x
Li W, Wang L-Y, Duan R-Y, Liu J-F, Gu J-D, Mu B-Z (2012) Microbial community characteristics of petroleum reservoir production water amended with n-alkanes and incubated under nitrate-, sulfate-reducing and methanogenic conditions. Int Biodeterior Biodegrad 69:87–96. doi:10.1016/j.ibiod.2012.01.005
Magot M, Ollivier B, Patel BKC (2000) Microbiology of petroleum reservoirs. Antonie Van Leeuwenhoek 77(2):103–116. doi:10.1023/A:1002434330514
Mbadinga S, Li K-P, Zhou L, Wang L-Y, Yang S-Z, Liu J-F, Gu J-D, Mu B-Z (2012) Analysis of alkane-dependent methanogenic community derived from production water of a high-temperature petroleum reservoir. Appl Microbiol Biotechnol 96(2):531–542. doi:10.1007/s00253-011-3828-8
Mbadinga SM, Wang L-Y, Zhou L, Liu J-F, Gu J-D, Mu B-Z (2011) Microbial communities involved in anaerobic degradation of alkanes. Int Biodeterior Biodegrad 65(1):1–13. doi:10.1016/j.ibiod.2010.11.009
McLellan SL, Huse SM, Mueller-Spitz SR, Andreishcheva EN, Sogin ML (2010) Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ Microbiol 12(2):378–392. doi:10.1111/j.1462-2920.2009.02075.x
Mukherjee S, Bardolui NK, Karim S, Patnaik VV, Nandy RK, Bag PK (2010) Isolation and characterization of a monoaromatic hydrocarbon-degrading bacterium, Pseudomonas aeruginosa from crude oil. J Environ Sci Health A 45(9):1048–1053. doi:10.1080/10934529.2010.486328
Navarro-Noya Y, Suárez-Arriaga M, Rojas-Valdes A, Montoya-Ciriaco N, Gómez-Acata S, Fernández-Luqueño F, Dendooven L (2013) Pyrosequencing analysis of the bacterial community in drinking water wells. Microb Ecol 66(1):19–29. doi:10.1007/s00248-013-0222-3
Nazina TN, Shestakova NM, Pavlova NK, Tatarkin YV, Ivoilov VS, Khisametdinov MR, Sokolova DS, Babich TL, Tourova TP, Poltaraus AB, Belyaev SS, Ivanov MV (2013) Functional and phylogenetic microbial diversity in formation waters of a low-temperature carbonate petroleum reservoir. Int Biodeterior Biodegrad 81:71–81. doi:10.1016/j.ibiod.2012.07.008
Obuekwe CO, Al-Jadi ZK, Al-Saleh ES (2008) Comparative hydrocarbon utilization by hydrophobic and hydrophilic variants of Pseudomonas aeruginosa. J Appl Microbiol 105(6):1876–1887. doi:10.1111/j.1365-2672.2008.03887.x
Orphan VJ, Taylor LT, Hafenbradl D, Delong EF (2000) Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl Environ Microbiol 66(2):700–711. doi:10.1128/aem.66.2.700-711.2000
Parkes J (1999) Cracking anaerobic bacteria. Nature 401(6750):217–218. doi:10.1038/45686
Pham VD, Hnatow LL, Zhang S, Fallon RD, Jackson SC, Tomb J-F, DeLong EF, Keeler SJ (2009) Characterizing microbial diversity in production water from an Alaskan mesothermic petroleum reservoir with two independent molecular methods. Environ Microbiol 11(1):176–187. doi:10.1111/j.1462-2920.2008.01751.x
Puech V, Chami M, Lemassu A, Lanéelle M-A, Schiffler B, Gounon P, Bayan N, Benz R, Daffé M (2001) Structure of the cell envelope of Corynebacteria: importance of the non-covalently bound lipids in the formation of the cell wall permeability barrier and fracture plane. Microbiol 147(5):1365–1382. doi:10.1073/pnas.1112572108
Ren H-Y, Zhang X-J, Z-y S, Rupert W, Gao G-J, S-x G, Zhao L-P (2011) Comparison of microbial community compositions of injection and production well samples in a long-term water-flooded petroleum reservoir. PLoS ONE 6(8):e23258. doi:10.1371/journal.pone.0023258
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541. doi:10.1128/aem.01541-09
Sen R (2008) Biotechnology in petroleum recovery: the microbial EOR. Prog Energy Combust Sci 34(6):714–724. doi:10.1016/j.pecs.2008.05.001
Singleton DR, Guzmán Ramirez L, Aitken MD (2009) Characterization of a polycyclic aromatic hydrocarbon degradation gene cluster in a phenanthrene-degrading Acidovorax strain. Appl Environ Microbiol 75(9):2613–2620. doi:10.1128/aem.01955-08
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. PNAS 103(32):12115–12120. doi:10.1073/pnas.0605127103
Suflita JM, Davidova IA, Gieg LM, Nanny M, Prince RC (2004) Anaerobic hydrocarbon biodegradation and the prospects for microbial enhanced energy production. In: Rafael V-D, Rodolfo Q-R (eds) Studies in surface science and catalysis. Elsevier, New York, pp 283–305
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. doi:10.1093/molbev/msr121
Tang Y-Q, Li Y, Zhao J-Y, Chi C-Q, Huang L-X, Dong H-P, Wu X-L (2012) Microbial communities in long-term, water-flooded petroleum reservoirs with different temperatures in the huabei oilfield, China. PLoS ONE 7(3):e33535. doi:10.1371/journal.pone.0033535
Thamdrup B, Rosselló-Mora R, Amann R (2000) Microbial manganese and sulfate reduction in black sea shelf sediments. Appl Environ Microbiol 66(7):2888–2897. doi:10.1128/aem.66.7.2888-2897.2000
Vasconcellos SP, Angolini CFF, García INS, Martins Dellagnezze B, Silva CC, Marsaioli AJ, Neto EVS, Oliveira VM (2010) Reprint of: Screening for hydrocarbon biodegraders in a metagenomic clone library derived from Brazilian petroleum reservoirs. Org Geochem 41(9):1067–1073. doi:10.1016/j.orggeochem.2010.08.003
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267. doi:10.1128/aem.00062-07
Wang L-Y, Mbadinga SM, Li H, Liu J-F, Yang S-Z Mu B-Z (2010) Anaerobic degradation of petroleum hydrocarbons and enlightenment of the prospects for microbial bio-gasification of residual oil. Microbiol 37(1):96–102 (in Chinese with English abstract)
Wang L-Y, Gao C-X, Mbadinga SM, Zhou L, Liu J-F, Gu J-D, Mu B-Z (2011) Characterization of an alkane-degrading methanogenic enrichment culture from production water of an oil reservoir after 274 days of incubation. Int Biodeterior Biodegrad 65(3):444–450. doi:10.1016/j.ibiod.2010.12.010
Wang L-Y, Dun R-Y, Liu J-F, Yang S-Z, Gu J-D, Mu B-Z (2012) Molecular analysis of the microbial community structures in water-flooding petroleum reservoirs with different temperatures. Biogeosciences 9(11):4645–4659. doi:10.5194/bg-9-4645-2012
Watanabe K, Watanabe K, Kodama Y, Syutsubo K, Harayama S (2000) Molecular characterization of bacterial populations in petroleum-contaminated groundwater discharged from underground crude oil storage cavities. Appl Environ Microbiol 66:4803–4809. doi:10.1128/AEM.66.11.4803-4809.2000
Watanabe K, Kodama Y, Kaku N (2002) Diversity and abundance of bacteria in an underground oil-storage cavity. BMC Microbiol 2:23. doi:10.1186/1471-2180-2-23
Webster G, Rinna J, Roussel EG, Fry JC, Weightman AJ, Parkes RJ (2010) Prokaryotic functional diversity in different biogeochemical depth zones in tidal sediments of the Severn Estuary, UK, revealed by stable-isotope probing. FEMS Microbiol Ecol 72(2):179–197. doi:10.1111/j.1574-6941.2010.00848.x
Zhang H, Kallimanis A, Koukkou A, Drainas C (2004) Isolation and characterization of novel bacteria degrading polycyclic aromatic hydrocarbons from polluted Greek soils. Appl Microbiol Biotechnol 65(1):124–131. doi:10.1007/s00253-004-1614-6
Zhang X, Yue S, Zhong H, Hua W, Chen R, Cao Y, Zhao L (2011) A diverse bacterial community in an anoxic quinoline-degrading bioreactor determined by using pyrosequencing and clone library analysis. Appl Microbiol Biotechnol 91(2):425–434. doi:10.1007/s00253-011-3296-1
Zhang F, She Y-H, Chai L-J, Banat IM, Zhang X-T, Shu F-C, Wang Z-L, Yu L-J, Hou D-J (2012) Microbial diversity in long-term water-flooded oil reservoirs with different in situ temperatures in China. Sci Rep 2:760. doi:10.1038/srep00760
Zhou L, Li K-P, Mbadinga SM, Yang S-Z, Gu J-D, Mu B-Z (2012) Analyses of n-alkanes degrading community dynamics of a high-temperature methanogenic consortium enriched from production water of a petroleum reservoir by a combination of molecular techniques. Ecotoxicology 21(6):1680–1691. doi:10.1007/s10646-012-0949-5
Acknowledgments
This work was supported by The National Science Foundation of China (no. 31200101), Shanghai Postdoctoral Scientific Program (no. 12R21412800), China Postdoctoral Science Foundation funded project (2013 T60426), and the NSFC/RGC Joint Research Fund (no. 41161160560, N_HKU 718/11). We thank Mr. Zhichao Zhou for constructing the heat map.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, LY., Ke, WJ., Sun, XB. et al. Comparison of bacterial community in aqueous and oil phases of water-flooded petroleum reservoirs using pyrosequencing and clone library approaches. Appl Microbiol Biotechnol 98, 4209–4221 (2014). https://doi.org/10.1007/s00253-013-5472-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-5472-y